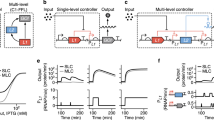
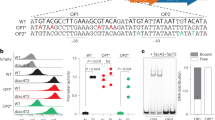
Abstract
Simple and predictable trans-acting regulatory tools are needed in the fields of synthetic biology and metabolic engineering to build complex genetic circuits and optimize the levels of native and heterologous gene products. Transcription activator−like effectors (TALEs) are bacterial virulence factors that have recently gained traction in biotechnology applications owing to their customizable DNA-binding specificity. In this work we expanded the versatility of these transcription factors to create an inducible TALE system by inserting tobacco-etch virus (TEV) protease recognition sites into the TALE backbone. The resulting engineered TALEs maintain transcriptional repression of their target genes in Escherichia coli, but are degraded after induction of the TEV protease, thereby promoting expression of the previously repressed target gene of interest. This TALE-TEV technology enables both repression and induction of plasmid or chromosomal target genes in a manner analogous to traditional repressor proteins but with the added flexibility of being operator-agnostic.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Kaern, M., Blake, W.J. & Collins, J.J. The engineering of gene regulatory networks. Annu. Rev. Biomed. Eng. 5, 179–206 (2003).
Brophy, J.A. & Voigt, C.A. Principles of genetic circuit design. Nat. Methods 11, 508–520 (2014).
Hsu, P.D., Lander, E.S. & Zhang, F. Development and applications of CRISPR-Cas9 for genome engineering. Cell 157, 1262–1278 (2014).
Sun, N. & Zhao, H. Transcription activator-like effector nucleases (TALENs): a highly efficient and versatile tool for genome editing. Biotechnol. Bioeng. 110, 1811–1821 (2013).
Boch, J. et al. Breaking the code of DNA binding specificity of TAL-type III effectors. Science 326, 1509–1512 (2009).
Jinek, M. et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012).
Citorik, R.J., Mimee, M. & Lu, T.K. Sequence-specific antimicrobials using efficiently delivered RNA-guided nucleases. Nat. Biotechnol. 32, 1141–1145 (2014).
Bikard, D. et al. Exploiting CRISPR-Cas nucleases to produce sequence-specific antimicrobials. Nat. Biotechnol. 32, 1146–1150 (2014).
Miyanari, Y., Ziegler-Birling, C. & Torres-Padilla, M.E. Live visualization of chromatin dynamics with fluorescent TALEs. Nat. Struct. Mol. Biol. 20, 1321–1324 (2013).
Chen, B. et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell 155, 1479–1491 (2013).
Qi, L.S. et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152, 1173–1183 (2013).
Politz, M.C., Copeland, M.F. & Pfleger, B.F. Artificial repressors for controlling gene expression in bacteria. Chem. Commun. (Camb.) 49, 4325–4327 (2013).
Nielsen, A.A. & Voigt, C.A. Multi-input CRISPR/Cas genetic circuits that interface host regulatory networks. Mol. Syst. Biol. 10, 763 (2014).
Gilbert, L.A. et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154, 442–451 (2013).
Perez-Pinera, P. et al. Synergistic and tunable human gene activation by combinations of synthetic transcription factors. Nat. Methods 10, 239–242 (2013).
Boissel, S. et al. megaTALs: a rare-cleaving nuclease architecture for therapeutic genome engineering. Nucleic Acids Res. 42, 2591–2601 (2014).
Li, Y., Moore, R., Guinn, M. & Bleris, L. Transcription activator-like effector hybrids for conditional control and rewiring of chromosomal transgene expression. Sci. Rep. 2, 897 (2012).
Konermann, S. et al. Optical control of mammalian endogenous transcription and epigenetic states. Nature 500, 472–476 (2013).
Mercer, A.C., Gaj, T., Sirk, S.J., Lamb, B.M. & Barbas III, C.F. Regulation of endogenous human gene expression by ligand-inducible TALE transcription factors. ACS Synth. Biol. 3, 723–730 (2014).
Lucast, L.J., Batey, R.T. & Doudna, J.A. Large-scale purification of a stable form of recombinant tobacco etch virus protease. Biotechniques 30, 544–546 (2001).
Bertrand, K.P., Postle, K., Wray, L.V. Jr. & Reznikoff, W.S. Overlapping divergent promoters control expression of Tn10 tetracycline resistance. Gene 23, 149–156 (1983).
Lienert, F. et al. Two- and three-input TALE-based AND logic computation in embryonic stem cells. Nucleic Acids Res. 41, 9967–9975 (2013).
Streubel, J., Blücher, C., Landgraf, A. & Boch, J. TAL effector RVD specificities and efficiencies. Nat. Biotechnol. 30, 593–595 (2012).
Mak, A.N., Bradley, P., Cernadas, R.A., Bogdanove, A.J. & Stoddard, B.L. The crystal structure of TAL effector PthXo1 bound to its DNA target. Science 335, 716–719 (2012).
Phan, J. et al. Structural basis for the substrate specificity of tobacco etch virus protease. J. Biol. Chem. 277, 50564–50572 (2002).
Dewey, D.L. & Work, E. Diaminopimelic acid decarboxylase and lysine: diaminopimelic acid decarboxylase. Nature 169, 533–534 (1952).
Kapust, R.B. et al. Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein Eng. 14, 993–1000 (2001).
Copeland, M.F., Politz, M.C. & Pfleger, B.F. Application of TALEs, CRISPR/Cas and sRNAs as trans-acting regulators in prokaryotes. Curr. Opin. Biotechnol. 29, 46–54 (2014).
de Lange, O., Binder, A. & Lahaye, T. From dead leaf, to new life: TAL effectors as tools for synthetic biology. Plant J. 78, 753–771 (2014).
Cuculis, L., Abil, Z., Zhao, H. & Schroeder, C.M. Direct observation of TALE protein dynamics reveals a two-state search mechanism. Nat. Commun. 6, 7277 (2015).
Kay, S., Hahn, S., Marois, E., Hause, G. & Bonas, U. A bacterial effector acts as a plant transcription factor and induces a cell size regulator. Science 318, 648–651 (2007).
Gao, H., Wu, X., Chai, J. & Han, Z. Crystal structure of a TALE protein reveals an extended N-terminal DNA binding region. Cell Res. 22, 1716–1720 (2012).
Meckler, J.F. et al. Quantitative analysis of TALE-DNA interactions suggests polarity effects. Nucleic Acids Res. 41, 4118–4128 (2013).
Garg, A., Lohmueller, J.J., Silver, P.A. & Armel, T.Z. Engineering synthetic TAL effectors with orthogonal target sites. Nucleic Acids Res. 40, 7584–7595 (2012).
Pérez-Quintero, A.L. et al. An improved method for TAL effectors DNA-binding sites prediction reveals functional convergence in TAL repertoires of Xanthomonas oryzae strains. PLoS One 8, e68464 (2013).
Mali, P. et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 31, 833–838 (2013).
Na, D. et al. Metabolic engineering of Escherichia coli using synthetic small regulatory RNAs. Nat. Biotechnol. 31, 170–174 (2013).
Esvelt, K.M. et al. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat. Methods 10, 1116–1121 (2013).
Zalatan, J.G. et al. Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds. Cell 160, 339–350 (2015).
Cameron, D.E. & Collins, J.J. Tunable protein degradation in bacteria. Nat. Biotechnol. 32, 1276–1281 (2014).
Guglielmi, L. & Martineau, P. Expression of single-chain Fv fragments in E. coli cytoplasm. Methods Mol. Biol. 562, 215–224 (2009).
Gibson, D.G. et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345 (2009).
Lynch, M.D. & Gill, R.T. Broad host range vectors for stable genomic library construction. Biotechnol. Bioeng. 94, 151–158 (2006).
Puigbò, P., Guzmán, E., Romeu, A. & Garcia-Vallvé, S. OPTIMIZER: a web server for optimizing the codon usage of DNA sequences. Nucleic Acids Res. 35, W126–W131 (2007).
Lee, T.S. et al. BglBrick vectors and datasheets: a synthetic biology platform for gene expression. J. Biol. Eng. 5, 12 (2011).
Huang, D., Holtz, W.J. & Maharbiz, M.M. A genetic bistable switch utilizing nonlinear protein degradation. J. Biol. Eng. 6, 9 (2012).
Stringer, A.M. et al. FRUIT, a scar-free system for targeted chromosomal mutagenesis, epitope tagging, and promoter replacement in Escherichia coli and Salmonella enterica. PLoS One 7, e44841 (2012).
Datsenko, K.A. & Wanner, B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97, 6640–6645 (2000).
Ellis, H.M., Yu, D., DiTizio, T. & Court, D.L. High efficiency mutagenesis, repair, and engineering of chromosomal DNA using single-stranded oligonucleotides. Proc. Natl. Acad. Sci. USA 98, 6742–6746 (2001).
Leveau, J.H. & Lindow, S.E. Predictive and interpretive simulation of green fluorescent protein expression in reporter bacteria. J. Bacteriol. 183, 6752–6762 (2001).
Pédelacq, J.D., Cabantous, S., Tran, T., Terwilliger, T.C. & Waldo, G.S. Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol. 24, 79–88 (2006).
Untergasser, A. et al. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 35, W71–W74 (2007).
Acknowledgements
This work was funded by the US National Science Foundation (NSF) (CBET-114678) and The William F. Vilas Trust. M.F.C. and M.C.P. were supported by NHGRI Genomic Sciences Training Program (T32 HG002760). A.L.M. was supported by NSF SEES fellowship (GEO-1215871). M.C.P. was supported by a US National Institutes of Health (NIH) Biotechnology Training Program (T32 GM008349). We thank D. Waugh for providing guidance on TEV protease variants, and J. Cameron for assistance with taking the photographs that appear in this paper.
Author information
Authors and Affiliations
Contributions
M.F.C. and B.F.P. conceived the study. M.F.C. and M.C.P. designed and performed the experiments with the following exceptions. A.L.M. proposed, and helped design and perform, the TEV Km mutant experiment. C.B.J. assisted with preliminary experimental work. M.F.C. and M.C.P. analyzed the data. M.F.C., M.C.P. and B.F.P. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Figures 1–12, Supplementary Tables 1–3 and Supplementary Note (PDF 5213 kb)
Rights and permissions
About this article
Cite this article
Copeland, M., Politz, M., Johnson, C. et al. A transcription activator–like effector (TALE) induction system mediated by proteolysis. Nat Chem Biol 12, 254–260 (2016). https://doi.org/10.1038/nchembio.2021
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.2021
This article is cited by
-
Directed evolution improves the catalytic efficiency of TEV protease
Nature Methods (2020)
-
Microbial CRISPRi and CRISPRa Systems for Metabolic Engineering
Biotechnology and Bioprocess Engineering (2019)
-
Designing cell function: assembly of synthetic gene circuits for cell biology applications
Nature Reviews Molecular Cell Biology (2018)
-
Engineered promoters enable constant gene expression at any copy number in bacteria
Nature Biotechnology (2018)
-
Recent advances in synthetic biology of cyanobacteria
Applied Microbiology and Biotechnology (2018)