Abstract
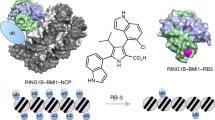
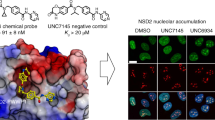
We report the design and characterization of UNC3866, a potent antagonist of the methyllysine (Kme) reading function of the Polycomb CBX and CDY families of chromodomains. Polycomb CBX proteins regulate gene expression by targeting Polycomb repressive complex 1 (PRC1) to sites of H3K27me3 via their chromodomains. UNC3866 binds the chromodomains of CBX4 and CBX7 most potently, with a Kd of ∼100 nM for each, and is 6- to 18-fold selective as compared to seven other CBX and CDY chromodomains while being highly selective over >250 other protein targets. X-ray crystallography revealed that UNC3866's interactions with the CBX chromodomains closely mimic those of the methylated H3 tail. UNC4195, a biotinylated derivative of UNC3866, was used to demonstrate that UNC3866 engages intact PRC1 and that EED incorporation into PRC1 is isoform dependent in PC3 prostate cancer cells. Finally, UNC3866 inhibits PC3 cell proliferation, consistent with the known ability of CBX7 overexpression to confer a growth advantage, whereas UNC4219, a methylated negative control compound, has negligible effects.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Wagner, T., Robaa, D., Sippl, W. & Jung, M. Mind the methyl: methyllysine binding proteins in epigenetic regulation. ChemMedChem 9, 466–483 (2014).
Arrowsmith, C.H., Bountra, C., Fish, P.V., Lee, K. & Schapira, M. Epigenetic protein families: a new frontier for drug discovery. Nat. Rev. Drug Discov. 11, 384–400 (2012).
Kaustov, L. et al. Recognition and specificity determinants of the human cbx chromodomains. J. Biol. Chem. 286, 521–529 (2011).
Cao, Q. et al. The central role of EED in the orchestration of polycomb group complexes. Nat. Commun. 5, 3127 (2014).
Vandamme, J., Volkel, P., Rosnoblet, C., Le Faou, P. & Angrand, P.O. Interaction proteomics analysis of polycomb proteins defines distinct PRC1 complexes in mammalian cells. Mol. Cell Proteomics 10, M110.002642 (2011).
Morey, L., Aloia, L., Cozzuto, L., Benitah, S.A. & Di Croce, L. RYBP and Cbx7 define specific biological functions of polycomb complexes in mouse embryonic stem cells. Cell Reports 3, 60–69 (2013).
Jin, B. et al. Linking DNA methyltransferases to epigenetic marks and nucleosome structure genome-wide in human tumor cells. Cell Rep. 2, 1411–1424 (2012).
Eskeland, R. et al. Ring1B compacts chromatin structure and represses gene expression independent of histone ubiquitination. Mol. Cell 38, 452–464 (2010).
Klauke, K. et al. Polycomb Cbx family members mediate the balance between haematopoietic stem cell self-renewal and differentiation. Nat. Cell Biol. 15, 353–362 (2013).
Bernard, D. et al. CBX7 controls the growth of normal and tumor-derived prostate cells by repressing the Ink4a/Arf locus. Oncogene 24, 5543–5551 (2005).
Forzati, F. et al. CBX7 is a tumor suppressor in mice and humans. J. Clin. Invest. 122, 612–623 (2012).
Shinjo, K. et al. Expression of chromobox homolog 7 (CBX7) is associated with poor prognosis in ovarian clear cell adenocarcinoma via TRAIL-induced apoptotic pathway regulation. Int. J. Cancer 135, 308–318 (2014).
Mansueto, G. et al. Identification of a new pathway for tumor progression: microRNA-181b up-regulation and CBX7 down-regulation by HMGA1 protein. Genes Cancer 1, 210–224 (2010).
Pallante, P. et al. Loss of the CBX7 gene expression correlates with a highly malignant phenotype in thyroid cancer. Cancer Res. 68, 6770–6778 (2008).
Pallante, P. et al. The loss of the CBX7 gene expression represents an adverse prognostic marker for survival of colon carcinoma patients. Eur. J. Cancer 46, 2304–2313 (2010).
Karamitopoulou, E. et al. Loss of the CBX7 protein expression correlates with a more aggressive phenotype in pancreatic cancer. Eur. J. Cancer 46, 1438–1444 (2010).
Zhang, X.W. et al. Oncogenic role of the chromobox protein CBX7 in gastric cancer. J. Exp. Clin. Cancer Res. 29, 114 (2010).
Gil, J., Bernard, D., Martínez, D. & Beach, D. Polycomb CBX7 has a unifying role in cellular lifespan. Nat. Cell Biol. 6, 67–72 (2004).
Scott, C.L. et al. Role of the chromobox protein CBX7 in lymphomagenesis. Proc. Natl. Acad. Sci. USA 104, 5389–5394 (2007).
Tan, J. et al. CBX8, a polycomb group protein, is essential for MLL-AF9-induced leukemogenesis. Cancer Cell 20, 563–575 (2011).
Wang, B. et al. Chromobox homolog 4 is correlated with prognosis and tumor cell growth in hepatocellular carcinoma. Ann. Surg. Oncol. 20 (suppl. 3): S684–S692 (2013).
Clermont, P.L. et al. Genotranscriptomic meta-analysis of the Polycomb gene CBX2 in human cancers: initial evidence of an oncogenic role. Br. J. Cancer 111, 1663–1672 (2014).
Sauvageau, M. & Sauvageau, G. Polycomb group genes: keeping stem cell activity in balance. PLoS Biol. 6, e113 (2008).
Konze, K.D. et al. An orally bioavailable chemical probe of the Lysine Methyltransferases EZH2 and EZH1. ACS Chem. Biol. 8, 1324–1334 (2013).
Ntziachristos, P. et al. Contrasting roles of histone 3 lysine 27 demethylases in acute lymphoblastic leukaemia. Nature 514, 513–517 (2014).
Hashizume, R. et al. Pharmacologic inhibition of histone demethylation as a therapy for pediatric brainstem glioma. Nat. Med. 20, 1394–1396 (2014).
Simhadri, C. et al. Chromodomain antagonists that target the polycomb-group methyllysine reader protein chromobox homolog 7 (CBX7). J. Med. Chem. 57, 2874–2883 (2014).
Ren, C. et al. Small-molecule modulators of methyl-lysine binding for the CBX7 chromodomain. Chem. Biol. 22, 161–168 (2015).
Frye, S.V. The art of the chemical probe. Nat. Chem. Biol. 6, 159–161 (2010).
Bunnage, M.E., Chekler, E.L. & Jones, L.H. Target validation using chemical probes. Nat. Chem. Biol. 9, 195–199 (2013).
Workman, P. & Collins, I. Probing the probes: fitness factors for small molecule tools. Chem. Biol. 17, 561–577 (2010).
Filippakopoulos, P. et al. Selective inhibition of BET bromodomains. Nature 468, 1067–1073 (2010).
James, L.I. et al. Discovery of a chemical probe for the L3MBTL3 methyllysine reader domain. Nat. Chem. Biol. 9, 184–191 (2013).
Hopkinson, R.J. et al. Is JmjC oxygenase catalysis limited to demethylation? Angew. Chem. Int. Edn. Engl. 52, 7709–7713 (2013).
Dickson, B.M. Approaching a parameter-free metadynamics. Phys. Rev. E 84, 037701 (2011).
Muñoz, V., Thompson, P.A., Hofrichter, J. & Eaton, W.A. Folding dynamics and mechanism of β-hairpin formation. Nature 390, 196–199 (1997).
Xiao, Y., Chen, C. & He, Y. Folding mechanism of β-hairpin trpzip2: heterogeneity, transition state and folding pathways. Int. J. Mol. Sci. 10, 2838–2848 (2009).
Fischle, W., Franz, H., Jacobs, S.A., Allis, C.D. & Khorasanizadeh, S. Specificity of the chromodomain Y chromosome family of chromodomains for lysine-methylated ARK(S/T) motifs. J. Biol. Chem. 283, 19626–19635 (2008).
Besnard, J. et al. Automated design of ligands to polypharmacological profiles. Nature 492, 215–220 (2012).
Kroeze, W.K. et al. PRESTO-Tango as an open-source resource for interrogation of the druggable human GPCRome. Nat. Struct. Mol. Biol. 22, 362–369 (2015).
Montgomery, N.D., Yee, D., Montgomery, S.A. & Magnuson, T. Molecular and functional mapping of EED motifs required for PRC2-dependent histone methylation. J. Mol. Biol. 374, 1145–1157 (2007).
Kuzmichev, A., Jenuwein, T., Tempst, P. & Reinberg, D. Different EZH2-containing complexes target methylation of histone H1 or nucleosomal histone H3. Mol. Cell 14, 183–193 (2004).
Ren, X., Vincenz, C. & Kerppola, T.K. Changes in the distributions and dynamics of polycomb repressive complexes during embryonic stem cell differentiation. Mol. Cell. Biol. 28, 2884–2895 (2008).
Martinez Molina, D. et al. Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science 341, 84–87 (2013).
Li, Q. et al. Polycomb CBX7 directly controls trimethylation of histone H3 at lysine 9 at the p16 locus. PLoS One 5, e13732 (2010).
Kong, Y., Cui, H., Ramkumar, C. & Zhang, H. Regulation of senescence in cancer and aging. J. Aging Res. 2011, 963172 (2011).
Jarrard, D.F. et al. Deletional, mutational, and methylation analyses of CDKN2 (p16/MTS1) in primary and metastatic prostate cancer. Genes Chromosom. Cancer 19, 90–96 (1997).
Yaqinuddin, A., Qureshi, S.A., Qazi, R. & Abbas, F. Down-regulation of DNMT3b in PC3 cells effects locus-specific DNA methylation, and represses cellular growth and migration. Cancer Cell Int. 8, 13 (2008).
Yaqinuddin, A., Qureshi, S.A., Qazi, R., Farooq, S. & Abbas, F. DNMT1 silencing affects locus specific DNA methylation and increases prostate cancer derived PC3 cell invasiveness. J. Urol. 182, 756–761 (2009).
Mirochnik, Y. et al. Androgen receptor drives cellular senescence. PLoS One 7, e31052 (2012).
Hess, B., Kutzner, C., van der Spoel, D. & Lindahl, E. GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 4, 435–447 (2008).
Tiwary, P. & Parrinello, M. From metadynamics to dynamics. Phys. Rev. Lett. 111, 230602 (2013).
Voter, A.F. Hyperdynamics: accelerated molecular dynamics of infrequent events. Phys. Rev. Lett. 78, 3908–3911 (1997).
Bussi, G., Donadio, D. & Parrinello, M. Canonical sampling through velocity rescaling. J. Chem. Phys. 126, 014101 (2007).
Wigle, T.J. et al. Screening for inhibitors of low-affinity epigenetic peptide-protein interactions: an AlphaScreen-based assay for antagonists of methyl-lysine binding proteins. J. Biomol. Screen. 15, 62–71 (2010).
Perfetti, M.T. et al. Identification of a fragment-like small molecule ligand for the methyl-lysine binding protein, 53BP1. ACS Chem. Biol. 10, 1072–1081 (2015).
Barsyte-Lovejoy, D. et al. (R)-PFI-2 is a potent and selective inhibitor of SETD7 methyltransferase activity in cells. Proc. Natl. Acad. Sci. USA 111, 12853–12858 (2014).
Kabsch, W. XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010).
Evans, P.R. & Murshudov, G.N. How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr. 69, 1204–1214 (2013).
McCoy, A.J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Acknowledgements
We thank K. Pearce (UNC Chapel Hill) for useful discussions and careful reading of the manuscript, O. Fedorov (SGC Oxford) for support with the bromodomain selectivity screening, A. Tumber (SGC Oxford) for support with the lysine demethylase selectivity screening and J.R. Walker for the review of the crystal structures. We thank G. Wang (UNC) for providing PHF1, PHF19, PHF23 and JARID1A protein constructs. We thank the labs of T. Magnuson (UNC) and D. Margolis (UNC) for providing EED and BMI-1 antibodies, respectively. We also thank N. Sciaky (UNC) for help with cell counting using the high content imaging microscope. Results shown in this report are derived from work performed at Argonne National Laboratory, Structural Biology Center at the Advanced Photon Source, a US Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under contract no. DE-AC02-06CH11357. The research described here was supported by the National Institute of General Medical Sciences, US National Institutes of Health (NIH, grant R01GM100919), the Carolina Partnership and the University Cancer Research Fund, University of North Carolina at Chapel Hill, and the Welch Foundation (G-1847). The SGC is a registered charity (no. 1097737) that receives funds from AbbVie, Boehringer Ingelheim, the Canada Foundation for Innovation (CFI), the Canadian Institutes of Health Research (CIHR), Genome Canada, Ontario Genomics Institute Grant OGI-055, GlaxoSmithKline, Janssen, Lilly Canada, the Novartis Research Foundation, the Ontario Ministry of Economic Development and Innovation, Pfizer, Takeda and Wellcome Trust Grant 092809/Z/10/Z. M.T.B. directs the Protein Array Core, which is supported by the Cancer Prevention Research Institute of Texas (RP130432) and by the Centre for Environmental and Molecular Carcinogenesis at the MD Anderson Cancer Center.
Author information
Authors and Affiliations
Contributions
J.I.S. designed and synthesized all compounds and related analogs, performed ITC studies and pulldown studies, rendered structural images, assisted with AlphaScreen studies and performed all cell culture; B.M.D. performed adaptively biased molecular dynamics studies and assisted in compound design; N.C. performed CellTiter-Glo assays and cellular proliferation assays; Y.L., W.T. and S.Q. solved the X-ray crystal structures of UNC3866 with the various CBX proteins; J.L.N. and S.H.C. expressed purified proteins; K.G.H. performed pulldown studies; C.S. and K.B. performed protein array experiments; F.L. performed methyltransferase assays; X.-P.H. performed GPCR functional assays. B.M.B. performed and analyzed AlphaScreen assays; G.S. performed EED ITC experiments; J.I.S., B.D.M., S.G.P., J.M., B.L.R., M.V., P.J.B., M.T.B., C.H.A, L.I.J. and S.V.F. designed studies and discussed results; J.I.S., L.I.J. and S.V.F. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Figures 1–20, Supplementary Tables 1–15 and Synthetic Procedures (PDF 3958 kb)
Rights and permissions
About this article
Cite this article
Stuckey, J., Dickson, B., Cheng, N. et al. A cellular chemical probe targeting the chromodomains of Polycomb repressive complex 1. Nat Chem Biol 12, 180–187 (2016). https://doi.org/10.1038/nchembio.2007
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.2007
This article is cited by
-
Fighting PRC1 via the RING
Nature Chemical Biology (2021)
-
Inhibiting CBX4 efficiently protects hepatocellular carcinoma cells against sorafenib resistance
British Journal of Cancer (2021)
-
Canonical PRC1 controls sequence-independent propagation of Polycomb-mediated gene silencing
Nature Communications (2019)
-
Targeting non-bromodomain chromatin readers
Nature Structural & Molecular Biology (2019)
-
C10ORF12 modulates PRC2 histone methyltransferase activity and H3K27me3 levels
Acta Pharmacologica Sinica (2019)