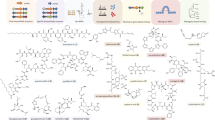
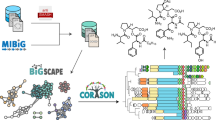
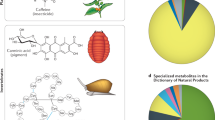
Abstract
Starting with the earliest Streptomyces genome sequences, the promise of natural product genome mining has been captivating: genomics and bioinformatics would transform compound discovery from an ad hoc pursuit to a high-throughput endeavor. Until recently, however, genome mining has advanced natural product discovery only modestly. Here, we argue that the development of algorithms to mine the continuously increasing amounts of (meta)genomic data will enable the promise of genome mining to be realized. We review computational strategies that have been developed to identify biosynthetic gene clusters in genome sequences and predict the chemical structures of their products. We then discuss networking strategies that can systematize large volumes of genetic and chemical data and connect genomic information to metabolomic and phenotypic data. Finally, we provide a vision of what natural product discovery might look like in the future, specifically considering longstanding questions in microbial ecology regarding the roles of metabolites in interspecies interactions.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Bentley, S.D. et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3 (2). Nature 417, 141–147 (2002).
Ikeda, H. et al. Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat. Biotechnol. 21, 526–531 (2003).
Medema, M.H., Breitling, R., Bovenberg, R. & Takano, E. Exploiting plug-and-play synthetic biology for drug discovery and production in microorganisms. Nat. Rev. Microbiol. 9, 131–137 (2011).
Bouslimani, A., Sanchez, L.M., Garg, N. & Dorrestein, P.C. Mass spectrometry of natural products: current, emerging and future technologies. Nat. Prod. Rep. 31, 718–729 (2014).
Krug, D. & Müller, R. Secondary metabolomics: the impact of mass spectrometry-based approaches on the discovery and characterization of microbial natural products. Nat. Prod. Rep. 31, 768–783 (2014).
Rappé, M.S. & Giovannoni, S.J. The uncultured microbial majority. Annu. Rev. Microbiol. 57, 369–394 (2003).
Epstein, S.S. The phenomenon of microbial uncultivability. Curr. Opin. Microbiol. 16, 636–642 (2013).
Streit, W.R. & Schmitz, R.A. Metagenomics—the key to the uncultured microbes. Curr. Opin. Microbiol. 7, 492–498 (2004).
Lasken, R.S. Genomic sequencing of uncultured microorganisms from single cells. Nat. Rev. Microbiol. 10, 631–640 (2012).
Klassen, J.L. & Currie, C.R. Gene fragmentation in bacterial draft genomes: extent, consequences and mitigation. BMC Genomics 13, 14 (2012).
Camacho, C. et al. BLAST+: architecture and applications. BMC Bioinformatics 10, 421 (2009).
Eddy, S.R. Accelerated profile HMM searches. PLOS Comput. Biol. 7, e1002195 (2011).
Weber, T. et al. CLUSEAN: a computer-based framework for the automated analysis of bacterial secondary metabolite biosynthetic gene clusters. J. Biotechnol. 140, 13–17 (2009).
Starcevic, A. et al. ClustScan: an integrated program package for the semi-automatic annotation of modular biosynthetic gene clusters and in silico prediction of novel chemical structures. Nucleic Acids Res. 36, 6882–6892 (2008).
Li, M.H., Ung, P.M., Zajkowski, J., Garneau-Tsodikova, S. & Sherman, D.H. Automated genome mining for natural products. BMC Bioinformatics 10, 185 (2009).
Khaldi, N. et al. SMURF: genomic mapping of fungal secondary metabolite clusters. Fungal Genet. Biol. 47, 736–741 (2010).
Medema, M.H. et al. antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 39, W339–W346 (2011).
Blin, K. et al. antiSMASH 2.0—a versatile platform for genome mining of secondary metabolite producers. Nucleic Acids Res. 41, W204–W212 (2013).
Eddy, S.R. Profile hidden Markov models. Bioinformatics 14, 755–763 (1998).
Fischbach, M.A. & Walsh, C.T. Antibiotics for emerging pathogens. Science 325, 1089–1093 (2009).
Rinke, C. et al. Insights into the phylogeny and coding potential of microbial dark matter. Nature 499, 431–437 (2013).
Cimermancic, P. et al. Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters. Cell 158, 412–421 (2014).
Punta, M. et al. The Pfam protein families database. Nucleic Acids Res. 40, D290–D301 (2012).
Pelzer, S., Wohlert, S.E. & Vente, A. Tool-box: tailoring enzymes for bio-combinatorial lead development and as markers for genome-based natural product lead discovery. Ernst Schering Res. Found. Workshop 51, 233–259 (2005).
Weng, J.-K. & Noel, J.P. The remarkable pliability and promiscuity of specialized metabolism. Cold Spring Harb. Symp. Quant. Biol. 77, 309–320 (2012).
Cruz-Morales, P. et al. Recapitulation of the evolution of biosynthetic gene clusters reveals hidden chemical diversity on bacterial genomes. bioRxiv doi:10.1101/020503.
Takeda, I., Umemura, M., Koike, H., Asai, K. & Machida, M. Motif-independent prediction of a secondary metabolism gene cluster using comparative genomics: application to sequenced genomes of Aspergillus and ten other filamentous fungal species. DNA Res. 21, 447–457 (2014).
Arnison, P.G. et al. Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 30, 108–160 (2013).
de Jong, A., van Hijum, S.A., Bijlsma, J.J., Kok, J. & Kuipers, O.P. BAGEL: a web-based bacteriocin genome mining tool. Nucleic Acids Res. 34, W273–W279 (2006).
de Jong, A., van Heel, A.J., Kok, J. & Kuipers, O.P. BAGEL2: mining for bacteriocins in genomic data. Nucleic Acids Res. 38, W647–W651 (2010).
Wilson, M.C. & Piel, J. Metagenomic approaches for exploiting uncultivated bacteria as a resource for novel biosynthetic enzymology. Chem. Biol. 20, 636–647 (2013).
Charlop-Powers, Z., Milshteyn, A. & Brady, S.F. Metagenomic small molecule discovery methods. Curr. Opin. Microbiol. 19, 70–75 (2014).
Reddy, B.V.B., Milshteyn, A., Charlop-Powers, Z. & Brady, S.F. eSNaPD: a versatile, web-based bioinformatics platform for surveying and mining natural product biosynthetic diversity from metagenomes. Chem. Biol. 21, 1023–1033 (2014).
Owen, J.G. et al. Mapping gene clusters within arrayed metagenomic libraries to expand the structural diversity of biomedically relevant natural products. Proc. Natl. Acad. Sci. USA 110, 11797–11802 (2013).
Ziemert, N. et al. The natural product domain seeker NaPDoS: a phylogeny based bioinformatic tool to classify secondary metabolite gene diversity. PLoS ONE 7, e34064 (2012).
Wu, D. et al. A phylogeny-driven genomic encyclopaedia of Bacteria and Archaea. Nature 462, 1056–1060 (2009).
Kampa, A. et al. Metagenomic natural product discovery in lichen provides evidence for a family of biosynthetic pathways in diverse symbioses. Proc. Natl. Acad. Sci. USA 110, E3129–E3137 (2013).
Kwan, J.C. et al. Genome streamlining and chemical defense in a coral reef symbiosis. Proc. Natl. Acad. Sci. USA 109, 20655–20660 (2012).
Boisvert, S., Raymond, F., Godzaridis, E., Laviolette, F. & Corbeil, J. Ray Meta: scalable de novo metagenome assembly and profiling. Genome Biol. 13, R122 (2012).
Howe, A.C. et al. Tackling soil diversity with the assembly of large, complex metagenomes. Proc. Natl. Acad. Sci. USA 111, 4904–4909 (2014).
Li, D., Liu, C.-M., Luo, R., Sadakane, K. & Lam, T.-W. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31, 1674–1676 (2015).
Albertsen, M. et al. Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat. Biotechnol. 31, 533–538 (2013).
Nielsen, H.B. et al. Identification and assembly of genomes and genetic elements in complex metagenomic samples without using reference genomes. Nat. Biotechnol. 32, 822–828 (2014).
Frasch, H.-J., Medema, M.H., Takano, E. & Breitling, R. Design-based re-engineering of biosynthetic gene clusters: plug-and-play in practice. Curr. Opin. Biotechnol. 24, 1144–1150 (2013).
Ziemert, N. et al. Diversity and evolution of secondary metabolism in the marine actinomycete genus Salinispora. Proc. Natl. Acad. Sci. USA 111, E1130–E1139 (2014).
Doroghazi, J.R. et al. A roadmap for natural product discovery based on large-scale genomics and metabolomics. Nat. Chem. Biol. 10, 963–968 (2014).
Yadav, G., Gokhale, R.S. & Mohanty, D. Computational approach for prediction of domain organization and substrate specificity of modular polyketide synthases. J. Mol. Biol. 328, 335–363 (2003).
Rausch, C., Weber, T., Kohlbacher, O., Wohlleben, W. & Huson, D.H. Specificity prediction of adenylation domains in nonribosomal peptide synthetases (NRPS) using transductive support vector machines (TSVMs). Nucleic Acids Res. 33, 5799–5808 (2005).
Minowa, Y., Araki, M. & Kanehisa, M. Comprehensive analysis of distinctive polyketide and nonribosomal peptide structural motifs encoded in microbial genomes. J. Mol. Biol. 368, 1500–1517 (2007).
Bachmann, B.O. & Ravel, J. Methods for in silico prediction of microbial polyketide and nonribosomal peptide biosynthetic pathways from DNA sequence data. Methods Enzymol. 458, 181–217 (2009).
Röttig, M. et al. NRPSpredictor2—a web server for predicting NRPS adenylation domain specificity. Nucleic Acids Res. 39, W362–W367 (2011).
Prieto, C., Garcia-Estrada, C., Lorenzana, D. & Martin, J.F. NRPSsp: non-ribosomal peptide synthase substrate predictor. Bioinformatics 28, 426–427 (2012).
Khayatt, B.I., Overmars, L., Siezen, R.J. & Francke, C. Classification of the adenylation and acyl-transferase activity of NRPS and PKS systems using ensembles of substrate specific hidden Markov models. PLoS ONE 8, e62136 (2013).
Baranašić, D. et al. Predicting substrate specificity of adenylation domains of nonribosomal peptide synthetases and other protein properties by latent semantic indexing. J. Ind. Microbiol. Biotechnol. 41, 461–467 (2014).
Blin, K., Kazempour, D., Wohlleben, W. & Weber, T. Improved lanthipeptide detection and prediction for antiSMASH. PLoS ONE 9, e89420 (2014).
Medema, M.H. et al. The Minimum Information about a Biosynthetic Gene cluster (MIBiG) specification. Nat. Chem. Biol. 11, 625–631 (2015).
Kersten, R.D. et al. A mass spectrometry-guided genome mining approach for natural product peptidogenomics. Nat. Chem. Biol. 7, 794–802 (2011).
Kersten, R.D. et al. Glycogenomics as a mass spectrometry-guided genome-mining method for microbial glycosylated molecules. Proc. Natl. Acad. Sci. USA 110, E4407–E4416 (2013).
Mohimani, H. et al. Automated genome mining of ribosomal peptide natural products. ACS Chem. Biol. 9, 1545–1551 (2014).
Mohimani, H. et al. NRPquest: coupling mass spectrometry and genome mining for nonribosomal peptide discovery. J. Nat. Prod. 77, 1902–1909 (2014).
Guthals, A., Watrous, J.D., Dorrestein, P.C. & Bandeira, N. The spectral networks paradigm in high throughput mass spectrometry. Mol. Biosyst. 8, 2535–2544 (2012).
Medema, M.H. et al. Pep2Path: automated mass spectrometry-guided genome mining of peptidic natural products. PLOS Comput. Biol. 10, e1003822 (2014).
Nguyen, D.D. et al. MS/MS networking guided analysis of molecule and gene cluster families. Proc. Natl. Acad. Sci. USA 110, E2611–E2620 (2013).
Schulze, C.J. et al. 'Function-first' lead discovery: mode of action profiling of natural product libraries using image-based screening. Chem. Biol. 20, 285–295 (2013).
Potts, M.B. et al. Using functional signature ontology (FUSION) to identify mechanisms of action for natural products. Sci. Signal. 6, ra90 (2013).
Yilmaz, P. et al. Minimum information about a marker gene sequence (MIMARKS) and minimum information about any (x) sequence (MIxS) specifications. Nat. Biotechnol. 29, 415–420 (2011).
Poulsen, M., Oh, D.-C., Clardy, J. & Currie, C.R. Chemical analyses of wasp-associated streptomyces bacteria reveal a prolific potential for natural products discovery. PLoS ONE 6, e16763 (2011).
Piel, J. et al. Exploring the chemistry of uncultivated bacterial symbionts: antitumor polyketides of the pederin family. J. Nat. Prod. 68, 472–479 (2005).
Yu, T.-W. et al. The biosynthetic gene cluster of the maytansinoid antitumor agent ansamitocin from Actinosynnema pretiosum. Proc. Natl. Acad. Sci. USA 99, 7968–7973 (2002).
Cardenas, M.E. et al. Antifungal activities of antineoplastic agents: Saccharomyces cerevisiae as a model system to study drug action. Clin. Microbiol. Rev. 12, 583–611 (1999).
Wilson, M.C. et al. An environmental bacterial taxon with a large and distinct metabolic repertoire. Nature 506, 58–62 (2014).
Crawford, J.M. & Clardy, J. Bacterial symbionts and natural products. Chem. Commun. (Camb.) 47, 7559–7566 (2011).
Bode, H.B. Entomopathogenic bacteria as a source of secondary metabolites. Curr. Opin. Chem. Biol. 13, 224–230 (2009).
van Heel, A.J., de Jong, A., Montalbán-López, M., Kok, J. & Kuipers, O.P. BAGEL3: Automated identification of genes encoding bacteriocins and (non-) bactericidal posttranslationally modified peptides. Nucleic Acids Res. 41, W448–W453 (2013).
Anand, S. et al. SBSPKS: structure based sequence analysis of polyketide synthases. Nucleic Acids Res. 8, W487–W496 (2010).
Medema, M.H., Takano, E. & Breitling, R. Detecting sequence homology at the gene cluster level with MultiGeneBlast. Mol. Biol. Evol. 30, 1218–1223 (2013).
Mohimani, H. et al. Cycloquest: identification of cyclopeptides via database search of their mass spectra against genome databases. J. Proteome Res. 10, 4505–4512 (2011).
Hadjithomas, M. et al. IMG-ABC: a knowledge base to fuel discovery of biosynthetic gene clusters and novel secondary metabolites. mBio. 6, e00932–15 (2015).
Ichikawa, N. et al. DoBISCUIT: a database of secondary metabolite biosynthetic gene clusters. Nucleic Acids Res. 41, D408–D414 (2013).
Conway, K.R. & Boddy, C.N. ClusterMine360: a database of microbial PKS/NRPS biosynthesis. Nucleic Acids Res. 41, D402–D407 (2013).
Diminic, J. et al. Databases of the thiotemplate modular systems (CSDB) and their in silico recombinants (r-CSDB). J. Ind. Microbiol. Biotechnol. 40, 653–659 (2013).
Tae, H., Sohng, J.K. & Park, K. MapsiDB: an integrated web database for type I polyketide synthases. Bioprocess Biosyst. Eng. 32, 723–727 (2009).
Hastings, J. et al. The ChEBI reference database and ontology for biologically relevant chemistry: enhancements for 2013. Nucleic Acids Res. 41, D456–D463 (2013).
Bento, A.P. et al. The ChEMBL bioactivity database: an update. Nucleic Acids Res. 42, D1083–D1090 (2014).
Nakamura, Y. et al. KNApSAcK metabolite activity database for retrieving the relationships between metabolites and biological activities. Plant Cell Physiol. 55, e7 (2014).
Wang, Y. et al. PubChem: a public information system for analyzing bioactivities of small molecules. Nucleic Acids Res. 37, W623–W633 (2009).
Pence, H.E. & Williams, A. ChemSpider: an online chemical information resource. J. Chem. Educ. 87, 1123–1124 (2010).
Caboche, S. et al. NORINE: a database of nonribosomal peptides. Nucleic Acids Res. 36, D326–D331 (2008).
Lucas, X. et al. StreptomeDB: a resource for natural compounds isolated from Streptomyces species. Nucleic Acids Res. 41, D1130–D1136 (2013).
Harborne, J.B. Dictionary of natural products. http://dnp.chemnetbase.com (Taylor & Francis, 2015).
Weber, T. In silico tools for the analysis of antibiotic biosynthetic pathways. Int. J. Med. Microbiol. 304, 230–235 (2014).
Boddy, C.N. Bioinformatics tools for genome mining of polyketide and non-ribosomal peptides. J. Ind. Microbiol. Biotechnol. 41, 443–450 (2014).
Kurtz, S. et al. Versatile and open software for comparing large genomes. Genome Biol. 5, R12 (2004).
Lin, K., Zhu, L. & Zhang, D.Y. An initial strategy for comparing proteins at the domain architecture level. Bioinformatics 22, 2081–2086 (2006).
Acknowledgements
We are indebted to P. Cimermancic, M. Donia and members of the Fischbach group for helpful conversations. This work was supported by a Rubicon grant of the Netherlands Organization for Scientific Research (NWO; Rubicon 825.13.001) to M.H.M. and by grants from the W.M. Keck Foundation (M.A.F.), the David and Lucile Packard Foundation (M.A.F.), the Glenn Foundation (M.A.F.), the Burroughs Wellcome Fund Investigators in the Pathogenesis of Infectious Disease program (M.A.F.), the Program for Breakthrough Biomedical Research (M.A.F.), US Defense Advanced Research Projects Agency (DARPA) award HR0011-12-C-0067 (M.A.F.) and US National Institutes of Health grants OD007290, AI101018, GM081879 and DK101674 (M.A.F.).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
M.A.F. is on the scientific advisory boards of NGM Biopharmaceuticals and Warp Drive Bio.
Rights and permissions
About this article
Cite this article
Medema, M., Fischbach, M. Computational approaches to natural product discovery. Nat Chem Biol 11, 639–648 (2015). https://doi.org/10.1038/nchembio.1884
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.1884
This article is cited by
-
CAGECAT: The CompArative GEne Cluster Analysis Toolbox for rapid search and visualisation of homologous gene clusters
BMC Bioinformatics (2023)
-
gutSMASH predicts specialized primary metabolic pathways from the human gut microbiota
Nature Biotechnology (2023)
-
Whole genome sequencing and analysis of multiple isolates of Ceratocystis destructans, the causal agent of Ceratocystis canker of almond in California
Scientific Reports (2023)
-
Ecological realism and rigor in the study of plant-plant allelopathic interactions
Plant and Soil (2023)
-
A Multi-Label Learning Framework for Predicting Chemical Classes and Biological Activities of Natural Products from Biosynthetic Gene Clusters
Journal of Chemical Ecology (2023)