Abstract
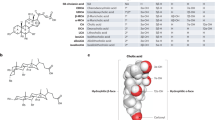
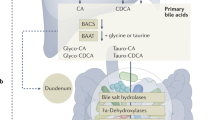
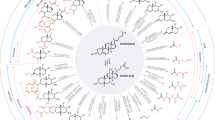
The gut bile acid pool is millimolar in concentration, varies widely in composition among individuals and is linked to metabolic disease and cancer. Although these molecules are derived almost exclusively from the microbiota, remarkably little is known about which bacterial species and genes are responsible for their biosynthesis. Here we report a biosynthetic pathway for the second most abundant class in the gut, 3β-hydroxy(iso)-bile acids, whose levels exceed 300 μM in some humans and are absent in others. We show, for the first time, that iso–bile acids are produced by Ruminococcus gnavus, a far more abundant commensal than previously known producers, and that the iso–bile acid pathway detoxifies deoxycholic acid and thus favors the growth of the keystone genus Bacteroides. By revealing the biosynthetic genes for an abundant class of bile acids, our work sets the stage for predicting and rationally altering the composition of the bile acid pool.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Accession codes
References
Ridlon, J.M., Kang, D.J. & Hylemon, P.B. Bile salt biotransformations by human intestinal bacteria. J. Lipid Res. 47, 241–259 (2006).
Hamilton, J.P. et al. Human cecal bile acids: concentration and spectrum. Am. J. Physiol. Gastrointest. Liver Physiol. 293, G256–G263 (2007).
Macdonald, I.A., Bokkenheuser, V.D., Winter, J., McLernon, A.M. & Mosbach, E.H. Degradation of steroids in the human gut. J. Lipid Res. 24, 675–700 (1983).
Hofmann, A.F. et al. A proposed nomenclature for bile acids. J. Lipid Res. 33, 599–604 (1992).
Setchell, K.D.R., Lawson, A.M., Tanida, N. & Sjovall, J. General methods for the analysis of metabolic profiles of bile acids and related compounds in feces. J. Lipid Res. 24, 1085–1100 (1983).
Thomas, C., Pellicciari, R., Pruzanski, M., Auwerx, J. & Schoonjans, K. Targeting bile-acid signalling for metabolic diseases. Nat. Rev. Drug Discov. 7, 678–693 (2008).
Makishima, M. et al. Vitamin D receptor as an intestinal bile acid sensor. Science 296, 1313–1316 (2002).
Talukdar, S., Bhatnagar, S., Dridi, S. & Hillgartner, F.B. Chenodeoxycholic acid suppresses the activation of acetyl–coenzyme A carboxylase–alpha gene transcription by the liver X receptor agonist T0–901317. J. Lipid Res. 48, 2647–2663 (2007).
Buffie, C.G. et al. Precision microbiome reconstitution restores bile acid–mediated resistance to Clostridium difficile. Nature 517, 205–208 (2015).
Reddy, B.S., Narisawa., T., Weisenburger, J.H. & Wynder, E.L. Promoting effect of sodium deoxycholate on colon adenocarcinomas in germfree rats. J. Natl. Cancer Inst. 56, 441–442 (1976).
Narisawa, T., Magadia, N.E., Weisburger, J.H. & Wynder, E.L. Promoting effect of bile acids on colon carcinogenesis after intrarectal instillation of N-methyl-N′-nitro-N-nitrosoguanidine in rats. J. Natl. Cancer Inst. 53, 1093–1097 (1974).
Yoshimoto, S. et al. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature 499, 97–101 (2013).
Rafter, J.J. et al. Cellular toxicity of fecal water depends on diet. Am. J. Clin. Nutr. 45, 559–563 (1987).
Reddy, B.S., Weisenburger, J.H. & Wynder, E.L. Effects of high risk and low risk diets for colon carcinogenesis on fecal microflora and steroids in man. J. Nutr. 105, 878–884 (1975).
Im, E. & Martinez, J.D. Ursodeoxycholic acid (UDCA) can inhibit deoxycholic acid (DCA)-induced apoptosis via modulation of EGFR/Raf-1/ERK signaling in human colon cancer cells. J. Nutr. 134, 483–486 (2004).
Bachrach, W.H. & Hofmann, A.F. Ursodeoxycholic acid in the treatment of cholesterol cholelithiasis. Part I. Dig. Dis. Sci. 27, 737–761 (1982).
Bennett, M.J., McKnight, S.L. & Coleman, J.P. Cloning and characterization of the NAD-dependent 7α-hydroxysteroid dehydrogenase from Bacteroides fragilis. Curr. Microbiol. 47, 475–484 (2003).
Liu, L., Aigner, A. & Schmid, R.D. Identification, cloning, heterologous expression and characterization of a NADPH-dependent 7β-hydroxysteroid dehydrogenase from Collinsella aerofaciens. Appl. Microbiol. Biotechnol. 90, 127–135 (2011).
Baron, S.F., Franklund, C.V. & Hylemon, P.B. Cloning, sequencing and expression of the gene encoding for bile acid 7α-hydroxysteroid dehydrogenase from Eubacterium sp. strain VPI 12708. J. Bacteriol. 173, 4558–4569 (1991).
Coleman, J.P., Hudson, L.L. & Adams, M.J. Characterization and regulation of the NADP-linked 7α-hydroxysteroid dehydrogenase gene from Clostridium sordellii. J. Bacteriol. 176, 4865–4874 (1994).
Lee, J.Y. et al. Contribution of the 7β-hydroxysteroid dehydrogenase from Ruminococcus gnavus N53 to ursodeoxycholic acid formation in the human colon. J. Lipid Res. 54, 3062–3069 (2013).
Hirano, S. & Masuda, N. Transformation of bile acids by Eubacterium lentum. Appl. Environ. Microbiol. 42, 912–915 (1981).
Hirano, S., Masuda, N., Oda, H. & Mukai, H. Transformation of bile acids by Clostridium perfringens. Appl. Environ. Microbiol. 42, 394–399 (1981).
Macdonald, I.A. et al. Metabolism of primary bile acids by Clostridium perfringens. J. Steroid Biochem. 18, 97–104 (1983).
Hayakawa, S. Microbiological transformation of bile acids. Adv. Lipid Res. 11, 143–192 (1973).
Macdonald, I.A., Meier, E.C., Mahony, D.E. & Costain, G.A. 3α-, 7α-, and 12α-hydroxysteroid dehydrogenase activity from Clostridium perfringens. Biochim. Biophys. Acta 450, 142–153 (1976).
MacDonald, I.A., Jellett, J.F., Mahony, D.E. & Holdeman, L.V. Bile salt 3α- and 12α-hydroxysteroid dehydrogenases from Eubacterium lentum and related organisms. Appl. Environ. Microbiol. 37, 992–1000 (1979).
MacDonald, I.A., Mahony, D.E., Jellet, J.F. & Meier, C.E. NAD-dependent 3α- and 12α-hydroxysteroid dehydrogenase activities from Eubacterium lentum ATCC no. 25559. Biochim. Biophys. Acta 489, 466–476 (1977).
Edenharder, R., Pfutzner, A. & Hammann, R. Characterization of NAD-dependent 3α- and 3β-hydroxysteroid dehydrogenase and of NADP-dependent 7β-hydroxysteroid dehydrogenase from Peptostreptococcus productus. Biochim. Biophys. Acta 1004, 230–238 (1989).
Edenharder, R., Pfutzner, A. & Hammann, R. NADP-dependent 3β-, 7α- and 7β-hydroxysteroid dehydrogenase activities from a lecithinase-lipase–negative Clostridium species 25.11.c. Biochim. Biophys. Acta 1002, 37–44 (1989).
Akao, T., Akao, T., Hattori, M., Namba, T. & Kobashi, K. 3β-hydroxysteroid dehydrogenase of Ruminococcus sp. from human intestinal bacteria. J. Biochem. 99, 1425–1431 (1986).
Kraal, L., Abubucker, S., Kota, K., Fischbach, M.A. & Mitreva, M. The prevalence of species and strains in the human microbiome: a resource for experimental efforts. PLoS ONE 9, e97279 (2014).
Wells, J.E. & Hylemon, P.B. Identification and characterization of a bile acid 7α-dehydroxylation operon in Clostridium sp. strain TO-931, a highly active 7α-dehydroxylating strain isolated from human feces. Appl. Environ. Microbiol. 66, 1107–1113 (2000).
Mallonee, D.H., Lijewski, M.A. & Hylemon, P.B. Expression in Escherichia coli and characterization of a bile acid–inducible 3α-hydroxysteroid dehydrogenase from Eubacterium sp. strain VPI 12708. Curr. Microbiol. 30, 259–263 (1995).
Ridlon, J.M., Kang, D. & Hylemon, P.B. Isolation and characterization of a bile acid inducible 7α-dehydroxylating operon in Clostridium hylemonae TN271. Anaerobe 16, 137–146 (2010).
Begley, M., Gahan, C.G.M. & Hill, C. The interaction between bacteria and bile. FEMS Microbiol. Rev. 29, 625–651 (2005).
Matsuoka, K. & Moroi, Y. Micelle formation of sodium deoxycholate and sodium ursodeoxycholate (part 1). Biochim. Biophys. Acta 1580, 189–199 (2002).
Gómez Zavaglia, A., Kociubinski, G., Perez, P., Disalvo, E. & De Antoni, G. Effect of bile on the lipid composition and surface properties of bifidobacteria. J. Appl. Microbiol. 93, 794–799 (2002).
Noh, D.O. & Gilliland, S.E. Influence of bile on cellular integrity and β-galactosidase activity of Lactobacillus acidophilus. J. Dairy Sci. 76, 1253–1259 (1993).
Maurice, C.F., Haiser, H.J. & Turnbaugh, P.J. Xenobiotics shape the physiology and gene expression of the active human gut microbiome. Cell 152, 39–50 (2013).
Holdeman, L.V. & Moore, W.E.C. New genus, Coprococcus, twelve new species, and emended descriptions of four previously described species of bacteria from human feces. Int. J. Syst. Bacteriol. 24, 260–277 (1974).
Lawson, A.M. & Setchell, K.D.R. The Bile Acids: Chemistry, Physiology and Metabolism Vol. 4 (eds. Setchell, K.D.R., Kritchevsky, D. & Nair, P.P.) 167–267 (Plenum Press, New York, USA, 1988).
Edgar, R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004).
Guindon, S. et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321 (2010).
Letunic, I. & Bork, P. Interactive tree of life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. 39, W475–W478 (2011).
Stiefel, P., Schmidt-Emrich, S., Maniura-Weber, K. & Ren, Q. Critical aspects of using bacterial cell viability assays with the fluorophores SYTO9 and propidium iodide. BMC Microbiol. 15, 36 (2015).
Acknowledgements
We are grateful to P. Turnbaugh (UCSF) for helpful discussions, to A. Hofmann (UCSD) for helpful discussions and for providing an initial sample of isoDCA and to T. Iida (Nihon University) for providing us with an authentic sample of 12-epi-isoDCA. We thank P.O. de Montellano for use of his GC-MS, Y. Varma for background research and experimentation on bile acids, P. Cimermancic for help with the phylogenetic analysis and members of the Fischbach group for helpful discussions. This research was supported by an Investigators in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund (M.A.F.), a Glenn Award for Research in Biological Mechanisms of Aging (M.A.F.), a Medical Research Program grant from the W.M. Keck Foundation (M.A.F.), a Fellowship for Science and Engineering from the David and Lucile Packard Foundation (M.A.F.) and US National Institutes of Health grants OD007290 and GM081879 (M.A.F.).
Author information
Authors and Affiliations
Contributions
A.S.D. and M.A.F. conceived the project, designed the experiments and wrote the manuscript. A.S.D. performed the experiments.
Corresponding author
Ethics declarations
Competing interests
M.A.F. is on the scientific advisory board of NGM Biopharmaceuticals.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Figures 1–14 and Supplementary Tables 1–4. (PDF 14755 kb)
Rights and permissions
About this article
Cite this article
Devlin, A., Fischbach, M. A biosynthetic pathway for a prominent class of microbiota-derived bile acids. Nat Chem Biol 11, 685–690 (2015). https://doi.org/10.1038/nchembio.1864
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.1864
This article is cited by
-
Dihydromyricetin ameliorated nonalcoholic steatohepatitis in mice by regulating the composition of serous lipids, bile acids and ileal microflora
Lipids in Health and Disease (2023)
-
Bile salt hydrolases shape the bile acid landscape and restrict Clostridioides difficile growth in the murine gut
Nature Microbiology (2023)
-
Investigation of the gut microbiome, bile acid composition and host immunoinflammatory response in a model of azoxymethane-induced colon cancer at discrete timepoints
British Journal of Cancer (2023)
-
Bile acids and the gut microbiota: metabolic interactions and impacts on disease
Nature Reviews Microbiology (2023)
-
Connecting the Gut Microbiota and Neurodegenerative Diseases: the Role of Bile Acids
Molecular Neurobiology (2023)