Abstract
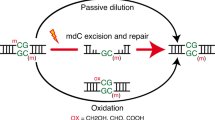
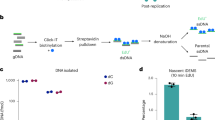
5-Formylcytosine (5fC) is a rare base found in mammalian DNA and thought to be involved in active DNA demethylation. Here, we show that developmental dynamics of 5fC levels in mouse DNA differ from those of 5-hydroxymethylcytosine (5hmC), and using stable isotope labeling in vivo, we show that 5fC can be a stable DNA modification. These results suggest that 5fC has functional roles in DNA that go beyond being a demethylation intermediate.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Ooi, S.K., O'Donnell, A.H. & Bestor, T.H. J. Cell Sci. 122, 2787–2791 (2009).
Goll, M.G. & Bestor, T.H. Annu. Rev. Biochem. 74, 481–514 (2005).
Kriaucionis, S. & Heintz, N. Science 324, 929–930 (2009).
Tahiliani, M. et al. Science 324, 930–935 (2009).
Globisch, D. et al. PLoS ONE 5, e15367 (2010).
Bachman, M. et al. Nat. Chem. 6, 1049–1055 (2014).
He, Y.F. et al. Science 333, 1303–1307 (2011).
Ito, S. et al. Science 333, 1300–1303 (2011).
Pfaffeneder, T. et al. Nat. Chem. Biol. 10, 574–581 (2014).
Song, C.X. & He, C. Trends Biochem. Sci. 38, 480–484 (2013).
Schiesser, S. et al. J. Am. Chem. Soc. 135, 14593–14599 (2013).
Neri, F. et al. Cell Reports 10, 674–683 (2015).
Cortázar, D. et al. Nature 470, 419–423 (2011).
Cortellino, S. et al. Cell 146, 67–79 (2011).
Hu, X. et al. Cell Stem Cell 14, 512–522 (2014).
Booth, M.J., Marsico, G., Bachman, M., Beraldi, D. & Balasubramanian, S. Nat. Chem. 6, 435–440 (2014).
Song, C.X. et al. Cell 153, 678–691 (2013).
Kraus, T.F., Guibourt, V. & Kretzschmar, H.A. J. Neural Transm. 10.1007/s00702-014-1346-4 (4 December 2014).
Iurlaro, M. et al. Genome Biol. 14, R119 (2013).
Spruijt, C.G. et al. Cell 152, 1146–1159 (2013).
Booth, M.J. et al. Science 336, 934–937 (2012).
Yu, M. et al. Cell 149, 1368–1380 (2012).
Lu, X. et al. Cell Res. 25, 386–389 (2015).
Raiber, E.A. et al. Nat. Struct. Mol. Biol. 22, 44–49 (2015).
Ying, Q.L. et al. Nature 453, 519–523 (2008).
Acknowledgements
We thank C. d'Santos and D. Oxley for their support with mass spectrometry and G. Xu for kindly providing TET-TKO mES cells. This work was supported by Cancer Research UK (C14303/A17197, S.B.), The Wellcome Trust (WT099232, S.B.; WT095645/Z/11/Z, W.R.) and the Biotechnology and Biological Sciences Research Council UK (BB/K010867/1, W.R.).
Author information
Authors and Affiliations
Contributions
M.B., S.U.-L. and S.B. conceived the study; S.U.-L., M.B., H.E.B. and M.I. performed experiments; M.B. and X.Y. carried out mass spectrometry and data analysis; S.B., A.M. and W.R. supervised the project; M.B. and S.B. wrote the manuscript with contributions from all authors.
Corresponding author
Ethics declarations
Competing interests
S.B. and W.R. are advisors and shareholders of Cambridge Epigenetix, Ltd.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Figures 1–9 and Supplementary Table 1 (PDF 922 kb)
Rights and permissions
About this article
Cite this article
Bachman, M., Uribe-Lewis, S., Yang, X. et al. 5-Formylcytosine can be a stable DNA modification in mammals. Nat Chem Biol 11, 555–557 (2015). https://doi.org/10.1038/nchembio.1848
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.1848
This article is cited by
-
Sperm chromatin accessibility’s involvement in the intergenerational effects of stress hormone receptor activation
Translational Psychiatry (2023)
-
Differential expression of m5C RNA methyltransferase genes NSUN6 and NSUN7 in Alzheimer’s disease and traumatic brain injury
Molecular Neurobiology (2023)
-
Isoform-specific and ubiquitination dependent recruitment of Tet1 to replicating heterochromatin modulates methylcytosine oxidation
Nature Communications (2022)
-
Unraveling the functional role of DNA demethylation at specific promoters by targeted steric blockage of DNA methyltransferase with CRISPR/dCas9
Nature Communications (2021)
-
Active turnover of DNA methylation during cell fate decisions
Nature Reviews Genetics (2021)