Abstract
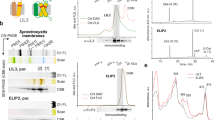
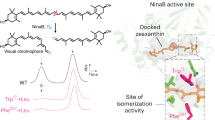
Plants synthesize carotenoids, which are essential for plant development and survival. These metabolites also serve as essential nutrients for human health. The biosynthetic pathway for all plant carotenoids occurs in chloroplasts and other plastids and requires 15-cis-ζ-carotene isomerase (Z-ISO). It was not known whether Z-ISO catalyzes isomerization alone or in combination with other enzymes. Here we show that Z-ISO is a bona fide enzyme and integral membrane protein. Z-ISO independently catalyzes the cis-trans isomerization of the 15-15′ carbon-carbon double bond in 9,15,9′-cis-ζ-carotene to produce the substrate required by the subsequent biosynthetic-pathway enzyme. We discovered that isomerization depends upon a ferrous heme b cofactor that undergoes redox-regulated ligand switching between the heme iron and alternate Z-ISO amino acid residues. Heme b–dependent isomerization of a large hydrophobic compound in a membrane was previously undescribed. As an isomerase, Z-ISO represents a new prototype for heme b proteins and potentially uses a new chemical mechanism.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
References
Moise, A.R., Al-Babili, S. & Wurtzel, E.T. Mechanistic aspects of carotenoid biosynthesis. Chem. Rev. 114, 164–193 (2014).
Wurtzel, E.T., Cuttriss, A. & Vallabhaneni, R. Maize provitamin A carotenoids, current resources and future metabolic engineering challenges. Front. Plant Sci. 3, 29 (2012).
Chen, Y., Li, F. & Wurtzel, E.T. Isolation and characterization of the Z-ISO gene encoding a missing component of carotenoid biosynthesis in plants. Plant Physiol. 153, 66–79 (2010).
Li, F., Murillo, C. & Wurtzel, E.T. Maize Y9 encodes a product essential for 15-cis zetacarotene isomerization. Plant Physiol. 144, 1181–1189 (2007).
Janick-Buckner, D., O'Neal, J., Joyce, E. & Buckner, B. Genetic and biochemical analysis of the y9 gene of maize, a carotenoid biosynthetic gene. Maydica 46, 41–46 (2001).
Tatusova, T.A. & Madden, T.L. Blast 2 sequences: a new tool for comparing protein and nucleotide sequences. FEMS Microbiol. Lett. 174, 247–250 (1999).
Jones, D.T. Improving the accuracy of transmembrane protein topology prediction using evolutionary information. Bioinformatics 23, 538–544 (2007).
Shumskaya, M. & Wurtzel, E.T. The carotenoid biosynthetic pathway: thinking in all dimensions. Plant Sci. 208, 58–63 (2013).
Ginalski, K., Elofsson, A., Fischer, D. & Rychlewski, L. 3D-Jury: a simple approach to improve protein structure predictions. Bioinformatics 19, 1015–1018 (2003).
Madej, M.G., Nasiri, H.R., Hilgendorff, N.S., Schwalbe, H. & Lancaster, C.R.D. Evidence for transmembrane proton transfer in a dihaem-containing membrane protein complex. EMBO J. 25, 4963–4970 (2006).
Vallat, B.K. et al. Building and assessing atomic models of proteins from structural templates: learning and benchmarks. Proteins 76, 930–945 (2009).
Thomas, P.E., Ryan, D. & Levin, W. An improved staining procedure for the detection of the peroxidase activity of cytochrome P-450 on sodium dodecyl sulfate polyacrylamide gels. Anal. Biochem. 75, 168–176 (1976).
Nygaard, T.K., Liu, M., McClure, M.J. & Lei, B. Identification and characterization of the heme-binding proteins SeShp and SeHtsA of Streptococcus equi subspecies equi. BMC Microbiol. 6, 82 (2006).
Owens, C.P., Du, J., Dawson, J.H. & Goulding, C.W. Characterization of heme ligation properties of Rv0203, a secreted heme binding protein involved in Mycobacterium tuberculosis heme uptake. Biochemistry 51, 1518–1531 (2012).
Boffi, A., Chiancone, E., Takahashi, S. & Rousseau, D.L. Stereochemistry of the Fe(II)- and Fe(III)-cyanide complexes of the homodimeric Scapharca inaequivalvis hemoglobin: a resonance Raman and FTIR study. Biochemistry 36, 4505–4509 (1997).
Tsai, A.L. et al. Heme coordination of prostaglandin H synthase. J. Biol. Chem. 268, 8554–8563 (1993).
Geng, J., Dornevil, K. & Liu, A. Chemical rescue of the distal histidine mutants of tryptophan 2,3-dioxygenase. J. Am. Chem. Soc. 134, 12209–12218 (2012).
Zoppellaro, G. et al. Review: studies of ferric heme proteins with highly anisotropic/highly axial low spin (S = 1/2) electron paramagnetic resonance signals with bis-histidine and histidine-methionine axial iron coordination. Biopolymers 91, 1064–1082 (2009).
Zhong, F., Lisi, G.P., Collins, D.P., Dawson, J.H. & Pletneva, E.V. Redox-dependent stability, protonation, and reactivity of cysteine-bound heme proteins. Proc. Natl. Acad. Sci. USA 111, E306–E315 (2014).
Smith, A.T. et al. Identification of Cys94 as the distal ligand to the Fe(III) heme in the transcriptional regulator RcoM-2 from Burkholderia xenovorans. J. Biol. Inorg. Chem. 17, 1071–1082 (2012).
Enemark, J.H. & Feltham, R.D. Principles of structure, bonding, and reactivity for metal nitrosyl complexes. Coord. Chem. Rev. 13, 339–406 (1974).
Yonetani, T., Yamamoto, H., Erman, J.E., Leigh, J.S. Jr. & Reed, G.H. Electromagnetic properties of hemoproteins. V. Optical and electron paramagnetic resonance characteristics of nitric oxide derivatives of metalloporphyrin-apohemoprotein complexes. J. Biol. Chem. 247, 2447–2455 (1972).
Sun, J., Wilks, A., Ortiz de Montellano, P.R. & Loehr, T.M. Resonance Raman and EPR spectroscopic studies on heme-heme oxygenase complexes. Biochemistry 32, 14151–14157 (1993).
Vickery, L., Nozawa, T. & Sauer, K. Magnetic circular dichroism studies of low-spin cytochromes: temperature dependence and effects of axial coordination on the spectra of cytochrome c and cytochrome b5. J. Am. Chem. Soc. 98, 351–357 (1976).
Vickery, L., Nozawa, T. & Sauer, K. Magnetic circular dichroism studies of myoglobin complexes: correlations with heme spin state and axial ligation. J. Am. Chem. Soc. 98, 343–350 (1976).
Sono, M., Dawson, J.H., Hall, K. & Hager, L.P. Ligand and halide binding properties of chloroperoxidase: peroxidase-type active site heme environment with cytochrome P-450 type endogenous axial ligand and spectroscopic properties. Biochemistry 25, 347–356 (1986).
Dawson, J.H., Andersson, L.A. & Sono, M. Spectroscopic investigations of ferric cytochrome P-450-CAM ligand complexes: identification of the ligand trans to cysteinate in the native enzyme. J. Biol. Chem. 257, 3606–3617 (1982).
Li, T., Bonkovsky, H.L. & Guo, J.T. Structural analysis of heme proteins: implications for design and prediction. BMC Struct. Biol. 11, 13 (2011).
Wouters, J. et al. Catalytic mechanism of Escherichia coli isopentenyl diphosphate isomerase involves Cys-67, Glu-116, and Tyr-104 as suggested by crystal structures of complexes with transition state analogues and irreversible inhibitors. J. Biol. Chem. 278, 11903–11908 (2003).
D'Angelo, P. et al. Unusual heme iron-lipid acyl chain coordination in Escherichia coli flavohemoglobin. Biophys. J. 86, 3882–3892 (2004).
Crabtree, R.H. in The Organometallic Chemistry of the Transition Metals 125–158 (John Wiley & Sons, 2005).
Pearson, R.G. Hard and soft acids and bases, HSAB, part II: underlying theories. J. Chem. Educ. 45, 643 (1968).
Heipieper, H.J., Neumann, G., Kabelitz, N., Kastner, M. & Richnow, H.H. Carbon isotope fractionation during cis-trans isomerization of unsaturated fatty acids in Pseudomonas putida. Appl. Microbiol. Biotechnol. 66, 285–290 (2004).
Williams, P.A. et al. Haem-ligand switching during catalysis in crystals of a nitrogen-cycle enzyme. Nature 389, 406–412 (1997).
Geng, J., Davis, I., Liu, F. & Liu, A. Bis-Fe(IV): nature's sniper for long-range oxidation. J. Biol. Inorg. Chem. 19, 1057–1067 (2014).
Richter, A.S. & Grimm, B. Thiol-based redox control of enzymes involved in the tetrapyrrole biosynthesis pathway. Front. Plant Sci. 4, 371 (2013).
Fanciullino, A.L., Bidel, L.P.R. & Urban, L. Carotenoid responses to environmental stimuli: integrating redox and carbon controls into a fruit model. Plant Cell Environ. 37, 273–289 (2014).
Davison, P.A. Overexpression of β-carotene hydroxylase enhances stress tolerance in Arabidopsis. Nature 418, 203–206 (2002).
Johnson, M.P. et al. Elevated zeaxanthin bound to oligomeric LHCII enhances the resistance of Arabidopsis to photooxidative stress by a lipid-protective, antioxidant mechanism. J. Biol. Chem. 282, 22605–22618 (2007).
Walter, M.H., Floss, D.S. & Strack, D. Apocarotenoids: hormones, mycorrhizal metabolites and aroma volatiles. Planta 232, 1–17 (2010).
Li, F., Vallabhaneni, R. & Wurtzel, E.T. PSY3, a new member of the phytoene synthase gene family conserved in the Poaceae and regulator of abiotic-stress-induced root carotenogenesis. Plant Physiol. 146, 1333–1345 (2008).
Gas, E., Flores-Perez, U., Sauret-Gueto, S. & Rodriguez-Concepcion, M. Hunting for plant nitric oxide synthase provides new evidence of a central role for plastids in nitric oxide metabolism. Plant Cell 21, 18–23 (2009).
Chang, H.-L., Hsu, Y.-T., Kang, C.-Y. & Lee, T.-M. Nitric oxide down-regulation of carotenoid synthesis and PSII activity in relation to very high light-induced singlet oxygen production and oxidative stress in Chlamydomonas reinhardtii. Plant Cell Physiol. 54, 1296–1315 (2013).
Khatsenko, O. Interactions between nitric oxide and cytochrome P-450 in the liver. Biochemistry (Mosc) 63, 833–839 (1998).
Cooper, C.E. Nitric oxide and iron proteins. Biochim. Biophys. Acta 1411, 290–309 (1999).
Munro, A., Girvan, H., McLean, K., Cheesman, M. & Leys, D. in Tetrapyrroles, Birth, Life and Death (eds. Warren, M. & Smith, A.) 160–183 (Landes Bioscience, Austin, Texas, USA, 2009).
Doyle, S.A. High-throughput cloning for proteomics research. Methods Mol. Biol. 310, 107–113 (2005).
Donnelly, M.I. et al. An expression vector tailored for large-scale, high-throughput purification of recombinant proteins. Protein Expr. Purif. 47, 446–454 (2006).
Seiler, C.Y. et al. DNASU plasmid and PSI:Biology-Materials repositories: resources to accelerate biological research. Nucleic Acids Res. 42, D1253–D1260 (2014).
Britton, G., Liaaen-Jensen, S. & Pfander, H. in Carotenoids: Spectroscopy Vol. 1B (eds. Britton, G., Liaaen-Jensen, S. & Pfander, H.) 13–62 (Birkhäuser, Basel, 1995).
Ter Horst, R. & Lolkema, J.S. Rapid screening of membrane topology of secondary transport proteins. Biochim Biophys Acta 1798, 672–680 (2010).
Shumskaya, M., Bradbury, L.M.T., Monaco, R.R. & Wurtzel, E.T. Plastid localization of the key carotenoid enzyme phytoene synthase is altered by isozyme, allelic variation, and activity. Plant Cell 24, 3725–3741 (2012).
Quinlan, R.F. et al. Synergistic interactions between carotene ring hydroxylases drive lutein formation in plant carotenoid biosynthesis. Plant Physiol. 160, 204–214 (2012).
Varanasi, L. & Hosler, J.P. Subunit III-depleted cytochrome c oxidase provides insight into the process of proton uptake by proteins. Biochim. Biophys. Acta 1817, 545–551 (2012).
Berry, E.A. & Trumpower, B.L. Simultaneous determination of hemes a, b, and c from pyridine hemochrome spectra. Anal. Biochem. 161, 1–15 (1987).
Thomas, P.E., Ryan, D. & Levin, W. An improved staining procedure for the detection of the peroxidase activity of cytochrome P-450 on sodium dodecyl sulfate polyacrylamide gels. Anal. Biochem. 75, 168–176 (1976).
Pond, A.E., Roach, M.P., Thomas, M.R., Boxer, S.G. & Dawson, J.H. The H93G myoglobin cavity mutant as a versatile template for modeling heme proteins: ferrous, ferric, and ferryl mixed-ligand complexes with imidazole in the cavity. Inorg. Chem. 39, 6061–6066 (2000).
Acknowledgements
We thank W. Hendrickson (Columbia University and New York Consortium on Membrane Protein Structure) for helpful discussions and use of the New York Structural Biology Center facilities. We thank M. Inouye (University of Medicine and Dentistry of New Jersey) for valuable advice on codon optimization and L. Bradbury (Lehman College and CUNY) for helpful discussions. E.T.W. was funded by the US National Institutes of Health (grant GM081160), CUNY and Lehman College. E.T.W. was also supported by travel funds from the Gordon Research Conferences to attend the Metals in Biology Gordon Research Conference, where E.T.W. was able to initiate collaborations with heme experts J.P. Hosler, A. Liu and J.H. Dawson through much-appreciated help from H. Gray (California Institute of Technology). The New York Consortium on Membrane protein structure (B.K. and J.D.L.) was supported through funds obtained from the US National Institutes of General Medical Sciences Protein Structure Initiative (PSI) program (grant GM095315). J.H.D. was funded by the US National Institutes of Health grant GM 26730; A.L. was funded by US National Institutes of Health grant R01GM108988 and the Georgia Research Alliance Distinguished Scholar Program. C.A.-D. was funded by The New Zealand Institute for Plant and Food Research Limited, New Zealand.
Author information
Authors and Affiliations
Contributions
Wet-laboratory experiments were performed by J.B. (cloning, protein expression and purification, enzyme assays, HPLC, carotenoid analyses, mutagenesis, heme assays and UV-vis spectroscopy) and B.K. (cloning and protein expression), J.D.L. (design of pNYCOMPS vector, cloning, expression and protein-purification protocols) and J.P.H. (ICP-OES, UV-vis spectroscopy, heme assays and CO binding). EPR experiments were performed by J.G. and A.L. MCD experiments were performed by A.M., M. Sono and J.H.D. Localization and import, including related gene cloning, was conducted by M. Shumskaya. J.B. and E.T.W. performed bioinformatic analyses. C.A.-D. prepared mutant Z-ISO fusion proteins while on a short-term sabbatical in the Wurtzel laboratory. All authors contributed to data analysis and to the writing and editing of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Tables 1–3 and Supplementary Figures 1–10 (PDF 1218 kb)
Rights and permissions
About this article
Cite this article
Beltrán, J., Kloss, B., Hosler, J. et al. Control of carotenoid biosynthesis through a heme-based cis-trans isomerase. Nat Chem Biol 11, 598–605 (2015). https://doi.org/10.1038/nchembio.1840
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.1840
This article is cited by
-
A foliar pigment-based bioassay for interrogating chloroplast signalling revealed that carotenoid isomerisation regulates chlorophyll abundance
Plant Methods (2022)
-
Carotenoids modulate kernel texture in maize by influencing amyloplast envelope integrity
Nature Communications (2020)
-
Environmental impacts on carotenoid metabolism in leaves
Plant Growth Regulation (2020)
-
A mutant allele of ζ-carotene isomerase (Z-ISO) is associated with the yellow pigmentation of the “Pinalate” sweet orange mutant and reveals new insights into its role in fruit carotenogenesis
BMC Plant Biology (2019)
-
Proteomic analysis of melatonin-mediated osmotic tolerance by improving energy metabolism and autophagy in wheat (Triticum aestivum L.)
Planta (2018)