Abstract
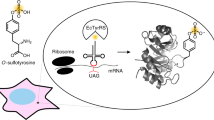
Serine phosphorylation is a key post-translational modification that regulates diverse biological processes. Powerful analytical methods have identified thousands of phosphorylation sites, but many of their functions remain to be deciphered. A key to understanding the function of protein phosphorylation is access to phosphorylated proteins, but this is often challenging or impossible. Here we evolve an orthogonal aminoacyl-tRNA synthetase/tRNACUA pair that directs the efficient incorporation of phosphoserine (pSer (1)) into recombinant proteins in Escherichia coli. Moreover, combining the orthogonal pair with a metabolically engineered E. coli enables the site-specific incorporation of a nonhydrolyzable analog of pSer. Our approach enables quantitative decoding of the amber stop codon as pSer, and we purify, with yields of several milligrams per liter of culture, proteins bearing biologically relevant phosphorylations that were previously challenging or impossible to access—including phosphorylated ubiquitin and the kinase Nek7, which is synthetically activated by a genetically encoded phosphorylation in its activation loop.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
References
Manning, G., Whyte, D.B., Martinez, R., Hunter, T. & Sudarsanam, S. The protein kinase complement of the human genome. Science 298, 1912–1934 (2002).
Olsen, J.V. et al. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127, 635–648 (2006).
Cohen, P. The structure and regulation of protein phosphatases. Annu. Rev. Biochem. 58, 453–508 (1989).
Ottesen, J.J., Huse, M., Sekedat, M.D. & Muir, T.W. Semisynthesis of phosphovariants of Smad2 reveals a substrate preference of the activated TβRI kinase. Biochemistry 43, 5698–5706 (2004).
Hejjaoui, M. et al. Elucidating the role of C-terminal post-translational modifications using protein semisynthesis strategies: α-synuclein phosphorylation at tyrosine 125. J. Am. Chem. Soc. 134, 5196–5210 (2012).
Sauerwald, A. et al. RNA-dependent cysteine biosynthesis in archaea. Science 307, 1969–1972 (2005).
Chin, J.W. Expanding and reprogramming the genetic code of cells and animals. Annu. Rev. Biochem. 83, 379–408 (2014).
Liu, C.C. & Schultz, P.G. Adding new chemistries to the genetic code. Annu. Rev. Biochem. 79, 413–444 (2010).
Fukunaga, R. & Yokoyama, S. Structural insights into the first step of RNA-dependent cysteine biosynthesis in archaea. Nat. Struct. Mol. Biol. 14, 272–279 (2007).
Park, H.S. et al. Expanding the genetic code of Escherichia coli with phosphoserine. Science 333, 1151–1154 (2011).
Lee, S. et al. A facile strategy for selective incorporation of phosphoserine into histones. Angew. Chem. Int. Edn Engl. 52, 5771–5775 (2013).
Eargle, J., Black, A.A., Sethi, A., Trabuco, L.G. & Luthey-Schulten, Z. Dynamics of recognition between tRNA and elongation factor Tu. J. Mol. Biol. 377, 1382–1405 (2008).
Heinemann, I.U. et al. Enhanced phosphoserine insertion during Escherichia coli protein synthesis via partial UAG codon reassignment and release factor 1 deletion. FEBS Lett. 586, 3716–3722 (2012).
Aerni, H.R., Shifman, M.A., Rogulina, S., O'Donoghue, P. & Rinehart, J. Revealing the amino acid composition of proteins within an expanded genetic code. Nucleic Acids Res. 43, e8 (2015).
Wang, K. et al. Optimized orthogonal translation of unnatural amino acids enables spontaneous protein double-labelling and FRET. Nat. Chem. 6, 393–403 (2014).
Chatterjee, A., Xiao, H. & Schultz, P.G. Evolution of multiple, mutually orthogonal prolyl-tRNA synthetase/tRNA pairs for unnatural amino acid mutagenesis in Escherichia coli. Proc. Natl. Acad. Sci. USA 109, 14841–14846 (2012).
Koyano, F. et al. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature 510, 162–166 (2014).
Kane, L.A. et al. PINK1 phosphorylates ubiquitin to activate parkin E3 ubiquitin ligase activity. J. Cell Biol. 205, 143–153 (2014).
Kazlauskaite, A. et al. Parkin is activated by PINK1-dependent phosphorylation of ubiquitin at Ser65. Biochem. J. 460, 127–139 (2014).
Pizer, L.I. The pathway and control of serine biosynthesis in Escherichia coli. J. Biol. Chem. 238, 3934–3944 (1963).
Zhang, C.M., Liu, C., Slater, S. & Hou, Y.M. Aminoacylation of tRNA with phosphoserine for synthesis of cysteinyl-tRNA(Cys). Nat. Struct. Mol. Biol. 15, 507–514 (2008).
Swanson, R. et al. Accuracy of in vivo aminoacylation requires proper balance of tRNA and aminoacyl-tRNA synthetase. Science 242, 1548–1551 (1988).
Wang, K., Neumann, H., Peak-Chew, S.Y. & Chin, J.W. Evolved orthogonal ribosomes enhance the efficiency of synthetic genetic code expansion. Nat. Biotechnol. 25, 770–777 (2007).
Tang, L. et al. Construction of “small-intelligent” focused mutagenesis libraries using well-designed combinatorial degenerate primers. Biotechniques 52, 149–158 (2012).
Nguyen, D.P., Garcia Alai, M.M., Kapadnis, P.B., Neumann, H. & Chin, J.W. Genetically encoding Nɛ-methyl-L-lysine in recombinant histones. J. Am. Chem. Soc. 131, 14194–14195 (2009).
Neumann, H., Wang, K., Davis, L., Garcia-Alai, M. & Chin, J.W. Encoding multiple unnatural amino acids via evolution of a quadruplet-decoding ribosome. Nature 464, 441–444 (2010).
Mukai, T. et al. Codon reassignment in the Escherichia coli genetic code. Nucleic Acids Res. 38, 8188–8195 (2010).
Lajoie, M.J. et al. Genomically recoded organisms expand biological functions. Science 342, 357–360 (2013).
Kondapalli, C. et al. PINK1 is activated by mitochondrial membrane potential depolarization and stimulates parkin E3 ligase activity by phosphorylating serine 65. Open Biol 2, 120080 (2012).
Valente, E.M. et al. Hereditary early-onset Parkinson's disease caused by mutations in PINK1. Science 304, 1158–1160 (2004).
Wauer, T. et al. Ubiquitin Ser65 phosphorylation affects ubiquitin structure, chain assembly and hydrolysis. EMBO J. 34, 307–325 (2015).
Belham, C. et al. A mitotic cascade of NIMA family kinases. Nercc1/Nek9 activates the Nek6 and Nek7 kinases. J. Biol. Chem. 278, 34897–34909 (2003).
O'Regan, L. & Fry, A.M. The Nek6 and Nek7 protein kinases are required for robust mitotic spindle formation and cytokinesis. Mol. Cell. Biol. 29, 3975–3990 (2009).
Richards, M.W. et al. An autoinhibitory tyrosine motif in the cell-cycle-regulated Nek7 kinase is released through binding of Nek9. Mol. Cell 36, 560–570 (2009).
Liu, F. et al. Serendipitous alkylation of a Plk1 ligand uncovers a new binding channel. Nat. Chem. Biol. 7, 595–601 (2011).
Klingberg, R. et al. Analysis of phosphorylation-dependent protein-protein interactions of histone H3. ACS Chem. Biol. 10, 138–145 (2015).
Neumann, H., Slusarczyk, A.L. & Chin, J.W. De novo generation of mutually orthogonal aminoacyl-tRNA synthetase/tRNA Pairs. J. Am. Chem. Soc. 132, 2142–2144 (2010).
Hohn, M.J., Park, H.S., O'Donoghue, P., Schnitzbauer, M. & Soll, D. Emergence of the universal genetic code imprinted in an RNA record. Proc. Natl. Acad. Sci. USA 103, 18095–18100 (2006).
Stemmer, W.P. & Morris, S.K. Enzymatic inverse PCR: a restriction site independent, single-fragment method for high-efficiency, site-directed mutagenesis. Biotechniques 13, 214–220 (1992).
Acknowledgements
We are grateful to the UK Medical Research Council Laboratory of Molecular Biology (MRC-LMB) Mass Spectrometry for extensive assistance. M. Richards (University of Leicester) for the Nek7 plasmid, T. Elliott (MRC-LMB), J. Madrzak (MRC-LMB) and M. Mahesh (MRC-LMB) for assistance. This work was supported by grants to J.W.C. from the UK Medical Research Council (U105181009 and UD99999908) and the European Research Council. M.M.K.M. is supported by the Wellcome Trust (101022/Z/13/Z), J. Macdonald Menzies Charitable Trust and Tenovus (Scotland). A.F.M. is supported by a Worldwide Cancer Research grant (13-0042) and R.B. by a Cancer Research UK Programme Award (C24461/A12772).
Author information
Authors and Affiliations
Contributions
D.T.R. and J.W.C. conceived the experimental strategy, analyzed the data and wrote the paper with input from other authors. D.T.R. performed all the selections, system characterization and most phosphoprotein expressions and purifications. S.M.H. and D.T.R. characterized the starting system. K.W., A.S., D.T.R. and N.H.-D. developed and characterized the expression system. T.H. and D.T.R. performed and analyzed the Nek7 experiments with guidance from A.M.F. and R.B. A.K. performed and analyzed the ubiquitin assays with guidance from M.M.K.M.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Tables 1–4 and Supplementary Figures 1–11 (PDF 12863 kb)
Rights and permissions
About this article
Cite this article
Rogerson, D., Sachdeva, A., Wang, K. et al. Efficient genetic encoding of phosphoserine and its nonhydrolyzable analog. Nat Chem Biol 11, 496–503 (2015). https://doi.org/10.1038/nchembio.1823
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.1823
This article is cited by
-
Virus-assisted directed evolution of enhanced suppressor tRNAs in mammalian cells
Nature Methods (2023)
-
Quintuply orthogonal pyrrolysyl-tRNA synthetase/tRNAPyl pairs
Nature Chemistry (2023)
-
Structural and functional consequences of NEDD8 phosphorylation
Nature Communications (2021)
-
Reprogramming the genetic code
Nature Reviews Genetics (2021)
-
Engineered triply orthogonal pyrrolysyl–tRNA synthetase/tRNA pairs enable the genetic encoding of three distinct non-canonical amino acids
Nature Chemistry (2020)