Abstract
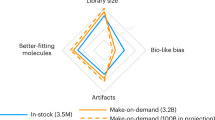
In lead discovery, libraries of 106 molecules are screened for biological activity. Given the over 1060 drug-like molecules thought possible, such screens might never succeed. The fact that they do, even occasionally, implies a biased selection of library molecules. We have developed a method to quantify the bias in screening libraries toward biogenic molecules. With this approach, we consider what is missing from screening libraries and how they can be optimized.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Wilhelm, S. et al. Discovery and development of sorafenib: a multikinase inhibitor for treating cancer. Nat. Rev. Drug Discov. 5, 835–844 (2006).
Spencer, R.W. High-throughput screening of historic collections: observations on file size, biological targets, and file diversity. Biotechnol. Bioeng. 61, 61–67 (1998).
Fox, S., Farr-Jones, S., Sopchak, L., Boggs, A. & Comley, J. High-throughput screening: searching for higher productivity. J. Biomol. Screen. 9, 354–358 (2004).
Macarron, R. Critical review of the role of HTS in drug discovery. Drug Discov. Today 11, 277–279 (2006).
Pereira, D.A. & Williams, J.A. Origin and evolution of high throughput screening. Br. J. Pharmacol. 152, 53–61 (2007).
Bohacek, R., McMartin, C. & Guida, W. The art and practice of structure-based drug design: a molecular modeling perspective. Med. Res. Rev. 16, 3–50 (1996).
Roth, B., Sheffler, D. & Kroeze, W. Magic shotguns versus magic bullets: selectively non-selective drugs for mood disorders and schizophrenia. Nat. Rev. Drug Discov. 3, 353–359 (2004).
Paolini, G., Shapland, R., van Hoorn, W., Mason, J. & Hopkins, A. Global mapping of pharmacological space. Nat. Biotechnol. 24, 805–815 (2006).
Yildirim, M., Goh, K.-I., Cusick, M., Barabasi, A.-L. & Vidal, M. Drug–target network. Nat. Biotechnol. 25, 1119–1126 (2007).
Martin, Y.C. Diverse viewpoints on computational aspects of molecular diversity. J. Comb. Chem. 3, 231–250 (2001).
Breinbauer, R., Vetter, I.R. & Waldmann, H. From protein domains to drug candidates—natural products as guiding principles in the design and synthesis of compound libraries. Angew. Chem. Int. Ed. 41, 2879–2890 (2002).
Koehn, F. & Carter, G. The evolving role of natural products in drug discovery. Nat. Rev. Drug Discov. 4, 206–220 (2005).
Arve, L., Voigt, T. & Waldmann, H. Charting biological and chemical space: PSSC and SCONP as guiding principles for the development of compound collections based on natural product scaffolds. QSAR Comb. Sci. 25, 449–456 (2006).
Ertl, P., Roggo, S. & Schuffenhauer, A. Natural product-likeness score and its application for prioritization of compound libraries. J. Chem. Inf. Model. 48, 68–74 (2008).
Gupta, S. Aires-de-Sousa, J. Comparing the chemical spaces of metabolites and available chemicals: models of metabolite-likeness. Mol. Divers. 11, 23–36 (2007).
Fink, T. & Reymond, J.L. Virtual exploration of the chemical universe up to 11 atoms of C, N, O, F: assembly of 26.4 million structures (110.9 million stereoisomers) and analysis for new ring systems, stereochemistry, physicochemical properties, compound classes, and drug discovery. J. Chem. Inf. Model. 47, 342–353 (2007).
Sadowski, J. & Kubinyi, H. A scoring scheme for discriminating between drugs and nondrugs. J. Med. Chem. 41, 3325–3329 (1998).
Good, A.C. & Hermsmeier, M.A. Measuring CAMD technique performance. 2. How “druglike” are drugs? Implications of random test set selection exemplified using druglikeness classification models. J. Chem. Inf. Model. 47, 110–114 (2007).
Glen, R.C. et al. Circular fingerprints: flexible molecular descriptors with applications from physical chemistry to ADME. IDrugs 9, 199–204 (2006).
Bemis, G.W. & Murcko, M.A. The properties of known drugs. 1. Molecular frameworks. J. Med. Chem. 39, 2887–2893 (1996).
Schreiber, S. Target-oriented and diversity-oriented organic synthesis in drug discovery. Science 287, 1964–1969 (2000).
Haggarty, S., Clemons, P., Wong, J. & Schreiber, S. Mapping chemical space using molecular descriptors and chemical genetics: deacetylase inhibitors. Comb. Chem. High Throughput Screen. 7, 669–676 (2004).
Shang, S. & Tan, D.S. Advancing chemistry and biology through diversity-oriented synthesis of natural product-like libraries. Curr. Opin. Chem. Biol. 9, 248–258 (2005).
Gregori-Puigjané, E. & Mestres, J. Coverage and bias in chemical library design. Curr. Opin. Chem. Biol. 12, 359–365 (2008).
Ertl, P., Jelfs, S., Mühlbacher, J., Schuffenhauer, A. & Selzer, P. Quest for the rings. In silico exploration of ring universe to identify novel bioactive heteroaromatic scaffolds. J. Med. Chem. 49, 4568–4573 (2006).
Wester, M.J. et al. Scaffold topologies. 2. Analysis of chemical databases. J. Chem. Inf. Model. 48, 1311–1324 (2008).
Wetzel, S., Schuffenhauer, A., Roggo, S., Ertl, P. & Waldmann, H. Cheminformatic analysis of natural products and their chemical space. Chimia 61, 355–360 (2007).
Fink, T., Bruggesser, H. & Reymond, J.L. Virtual exploration of the small-molecule chemical universe below 160 Daltons. Angew. Chem. Int. Ed. 44, 1504–1508 (2005).
Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 (2000).
Buckingham, J. Dictionary of Natural Products (Chapman & Hall/CRC, United Kingdom, 2008).
Irwin, J.J. & Shoichet, B.K. ZINC–a free database of commercially available compounds for virtual screening. J. Chem. Inf. Model. 45, 177–182 (2005).
Morgan, H.L. Generation of a unique description for chemical structures-a technique developed at Chemical Abstract Service. J. Chem. Doc. 5, 107–113 (1965).
Hert, J. et al. Comparison of topological descriptors for similarity-based virtual screening using multiple bioactive reference structures. Org. Biomol. Chem. 2, 3256–3266 (2004).
Koch, M. et al. Charting biologically relevant chemical space: a structural classification of natural products (SCONP). Proc. Natl. Acad. Sci. USA 102, 17272–17277 (2005).
Acknowledgements
This work was supported by US National Institutes of Health grant GM59957 to B.K.S. J.H. was supported by a Marie Curie fellowship from the 6th Framework Program of the European Commission; M.J.K. was supported by a US National Science Foundation graduate fellowship; C.L. was supported by a fellowship from the Max Kade Foundation.
Author information
Authors and Affiliations
Contributions
The project was conceived of by J.H. and B.K.S. J.H. undertook most of the calculations, with molecular proof checking by J.J.I. and C.L. and algorithmic assistance from M.J.K. J.H. and B.K.S. wrote the manuscript, which was read and commented on by the other authors.
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–4 and Supplementary Tables 1–3 (PDF 306 kb)
Rights and permissions
About this article
Cite this article
Hert, J., Irwin, J., Laggner, C. et al. Quantifying biogenic bias in screening libraries. Nat Chem Biol 5, 479–483 (2009). https://doi.org/10.1038/nchembio.180
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.180
This article is cited by
-
Combatting over-specialization bias in growing chemical databases
Journal of Cheminformatics (2023)
-
Modeling the expansion of virtual screening libraries
Nature Chemical Biology (2023)
-
A geometric deep learning approach to predict binding conformations of bioactive molecules
Nature Machine Intelligence (2021)
-
Marine dissolved organic matter: a vast and unexplored molecular space
Applied Microbiology and Biotechnology (2021)
-
De novo generation of hit-like molecules from gene expression signatures using artificial intelligence
Nature Communications (2020)