Abstract
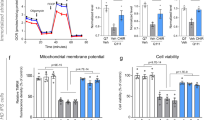
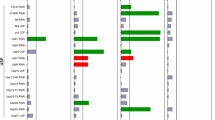
Huntington's disease (HD) is a currently incurable neurodegenerative condition caused by an abnormally expanded polyglutamine tract in huntingtin (HTT). We identified new modifiers of mutant HTT toxicity by performing a large-scale 'druggable genome' siRNA screen in human cultured cells, followed by hit validation in Drosophila. We focused on glutaminyl cyclase (QPCT), which had one of the strongest effects on mutant HTT-induced toxicity and aggregation in the cell-based siRNA screen and also rescued these phenotypes in Drosophila. We found that QPCT inhibition induced the levels of the molecular chaperone αB-crystallin and reduced the aggregation of diverse proteins. We generated new QPCT inhibitors using in silico methods followed by in vitro screening, which rescued the HD-related phenotypes in cell, Drosophila and zebrafish HD models. Our data reveal a new HD druggable target affecting mutant HTT aggregation and provide proof of principle for a discovery pipeline from druggable genome screen to drug development.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Imarisio, S. et al. Huntington's disease: from pathology and genetics to potential therapies. Biochem. J. 412, 191–209 (2008).
Zuccato, C., Valenza, M. & Cattaneo, E. Molecular mechanisms and potential therapeutical targets in Huntington's disease. Physiol. Rev. 90, 905–981 (2010).
A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. The Huntington's Disease Collaborative Research Group. Cell 72, 971–983 (1993).
Mangiarini, L. et al. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell 87, 493–506 (1996).
Hodgson, J.G. et al. A YAC mouse model for Huntington's disease with full-length mutant huntingtin, cytoplasmic toxicity, and selective striatal neurodegeneration. Neuron 23, 181–192 (1999).
Soto, C. & Estrada, L.D. Protein misfolding and neurodegeneration. Arch. Neurol. 65, 184–189 (2008).
Takahashi, T. et al. Soluble polyglutamine oligomers formed prior to inclusion body formation are cytotoxic. Hum. Mol. Genet. 17, 345–356 (2008).
Lajoie, P. & Snapp, E.L. Formation and toxicity of soluble polyglutamine oligomers in living cells. PLoS ONE 5, e15245 (2010).
Hartl, F.U., Bracher, A. & Hayer-Hartl, M. Molecular chaperones in protein folding and proteostasis. Nature 475, 324–332 (2011).
Sathasivam, K. et al. Aberrant splicing of HTT generates the pathogenic exon 1 protein in Huntington disease. Proc. Natl. Acad. Sci. USA 110, 2366–2370 (2013).
Bleicher, K.H., Böhm, H.-J., Müller, K. & Alanine, A.I. Hit and lead generation: beyond high-throughput screening. Nat. Rev. Drug Discov. 2, 369–378 (2003).
Li, M., Huang, Y., Ma, A.A.K., Lin, E. & Diamond, M.I. Y-27632 improves rotarod performance and reduces huntingtin levels in R6/2 mice. Neurobiol. Dis. 36, 413–420 (2009).
Marsh, J.L. et al. Expanded polyglutamine peptides alone are intrinsically cytotoxic and cause neurodegeneration in Drosophila. Hum. Mol. Genet. 9, 13–25 (2000).
Dietzl, G. et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448, 151–156 (2007).
Schilling, S. et al. Isolation and characterization of glutaminyl cyclases from Drosophila: evidence for enzyme forms with different subcellular localization. Biochemistry 46, 10921–10930 (2007).
Jackson, G.R. et al. Polyglutamine-expanded human huntingtin transgenes induce degeneration of Drosophila photoreceptor neurons. Neuron 21, 633–642 (1998).
Franceschini, N. & Kirschfeld, K. Pseudopupil phenomena in the compound eye of Drosophila. Kybernetik 9, 159–182 (1971).
Ravikumar, B. et al. Dynein mutations impair autophagic clearance of aggregate-prone proteins. Nat. Genet. 37, 771–776 (2005).
Zhang, S., Binari, R., Zhou, R. & Perrimon, N. A genomewide RNA interference screen for modifiers of aggregates formation by mutant Huntingtin in Drosophila. Genetics 184, 1165–1179 (2010).
Wyttenbach, A. et al. Effects of heat shock, heat shock protein 40 (HDJ-2), and proteasome inhibition on protein aggregation in cellular models of Huntington's disease. Proc. Natl. Acad. Sci. USA 97, 2898–2903 (2000).
Cynis, H. et al. Inhibition of glutaminyl cyclase alters pyroglutamate formation in mammalian cells. Biochim. Biophys. Acta 1764, 1618–1625 (2006).
Saido, T.C. Involvement of polyglutamine endolysis followed by pyroglutamate formation in the pathogenesis of triplet repeat/polyglutamine-expansion diseases. Med. Hypotheses 54, 427–429 (2000).
Onodera, O. et al. Oligomerization of expanded-polyglutamine domain fluorescent fusion proteins in cultured mammalian cells. Biochem. Biophys. Res. Commun. 238, 599–605 (1997).
Rankin, J., Wyttenbach, A. & Rubinsztein, D.C. Intracellular green fluorescent protein–polyalanine aggregates are associated with cell death. Biochem. J. 348, 15–19 (2000).
Luo, S., Mizuta, H. & Rubinsztein, D.C. p21-activated kinase 1 promotes soluble mutant huntingtin self-interaction and enhances toxicity. Hum. Mol. Genet. 17, 895–905 (2008).
Buchholz, M. et al. The first potent inhibitors for human glutaminyl cyclase: synthesis and structure-activity relationship. J. Med. Chem. 49, 664–677 (2006).
Schilling, S. et al. Glutaminyl cyclase inhibition attenuates pyroglutamate Aβ and Alzheimer's disease–like pathology. Nat. Med. 14, 1106–1111 (2008).
Schilling, S. et al. Continuous spectrometric assays for glutaminyl cyclase activity. Anal. Biochem. 303, 49–56 (2002).
Yoon, S. & Welsh, W.J. Identification of a minimal subset of receptor conformations for improved multiple conformation docking and two-step scoring. J. Chem. Inf. Comput. Sci. 44, 88–96 (2004).
Polgár, T. & Keserü, G.M. Ensemble docking into flexible active sites. Critical evaluation of FlexE against JNK-3 and β-secretase. J. Chem. Inf. Model. 46, 1795–1805 (2006).
Huang, K.-F., Liu, Y.-L., Cheng, W.-J., Ko, T.-P. & Wang, A.H.-J. Crystal structures of human glutaminyl cyclase, an enzyme responsible for protein N-terminal pyroglutamate formation. Proc. Natl. Acad. Sci. USA 102, 13117–13122 (2005).
Huang, K.-F. et al. Structures of human Golgi-resident glutaminyl cyclase and its complexes with inhibitors reveal a large loop movement upon inhibitor binding. J. Biol. Chem. 286, 12439–12449 (2011).
Ruiz-Carrillo, D. et al. Structures of glycosylated mammalian glutaminyl cyclases reveal conformational variability near the active center. Biochemistry 50, 6280–6288 (2011).
Jones, G., Willett, P. & Glen, R.C. Molecular recognition of receptor sites using a genetic algorithm with a description of desolvation. J. Mol. Biol. 245, 43–53 (1995).
Jones, G., Willett, P., Glen, R.C., Leach, A.R. & Taylor, R. Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 267, 727–748 (1997).
Verdonk, M.L., Cole, J.C., Hartshorn, M.J., Murray, C.W. & Taylor, R.D. Improved protein-ligand docking using GOLD. Proteins 52, 609–623 (2003).
Palm, K., Luthman, K., Ros, J., Grasjo, J. & Artursson, P. Effect of molecular charge on intestinal epithelial drug transport: pH-dependent transport of cationic drugs. J. Pharmacol. Exp. Ther. 291, 435–443 (1999).
Song, H. et al. Inhibition of glutaminyl cyclase ameliorates amyloid pathology in an animal model of Alzheimer's disease via the modulation of γ-secretase activity. J. Alzheimers Dis. 43, 797–807 (2014).
Robertson, A.L. et al. Small heat-shock proteins interact with a flanking domain to suppress polyglutamine aggregation. Proc. Natl. Acad. Sci. USA 107, 10424–10429 (2010).
Bilen, J. & Bonini, N.M. Drosophila as a model for human neurodegenerative disease. Annu. Rev. Genet. 39, 153–171 (2005).
Tue, N.T., Shimaji, K., Tanaka, N. & Yamaguchi, M. Effect of αB-crystallin on protein aggregation in Drosophila. J. Biomed. Biotechnol. 2012, 252049 (2012).
Williams, A. et al. Novel targets for Huntington's disease in an mTOR-independent autophagy pathway. Nat. Chem. Biol. 4, 295–305 (2008).
Lejeune, F.-X. et al. Large-scale functional RNAi screen in C. elegans identifies genes that regulate the dysfunction of mutant polyglutamine neurons. BMC Genomics 13, 91 (2012).
Miller, J.P. et al. Matrix metalloproteinases are modifiers of huntingtin proteolysis and toxicity in Huntington's disease. Neuron 67, 199–212 (2010).
Miller, J.P. et al. A genome-scale RNA-interference screen identifies RRAS signaling as a pathologic feature of Huntington's disease. PLoS Genet. 8, e1003042 (2012).
Yamanaka, T. et al. Large-scale RNA interference screening in mammalian cells identifies novel regulators of mutant huntingtin aggregation. PLoS ONE 9, e93891 (2014).
Waudby, C.A. et al. The interaction of αB-crystallin with mature α-synuclein amyloid fibrils inhibits their elongation. Biophys. J. 98, 843–851 (2010).
Raman, B. et al. αB-crystallin, a small heat-shock protein, prevents the amyloid fibril growth of an amyloid β-peptide and β2-microglobulin. Biochem. J. 392, 573–581 (2005).
Hochberg, G.K.A. et al. The structured core domain of αB-crystallin can prevent amyloid fibrillation and associated toxicity. Proc. Natl. Acad. Sci. USA 111, E1562–E1570 (2014).
Freeman, M. Reiterative use of the EGF receptor triggers differentiation of all cell types in the Drosophila eye. Cell 87, 651–660 (1996).
Narain, Y., Wyttenbach, A., Rankin, J., Furlong, R.A. & Rubinsztein, D.C. A molecular investigation of true dominance in Huntington's disease. J. Med. Genet. 36, 739–746 (1999).
D'Agostino, M. et al. The cytosolic chaperone α-crystallin B rescues folding and compartmentalization of misfolded multispan transmembrane proteins. J. Cell Sci. 126, 4160–4172 (2013).
Renna, M., Caporaso, M.G., Bonatti, S., Kaufman, R.J. & Remondelli, P. Regulation of ERGIC-53 gene transcription in response to endoplasmic reticulum stress. J. Biol. Chem. 282, 22499–22512 (2007).
Howarth, J.L. et al. Hsp40 molecules that target to the ubiquitin-proteasome system decrease inclusion formation in models of polyglutamine disease. Mol. Ther. 15, 1100–1105 (2007).
Westerfield, M. The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish (Danio rerio) (University of Oregon Press, 1995).
Kimmel, C.B., Ballard, W.W., Kimmel, S.R., Ullmann, B. & Schilling, T.F. Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253–310 (1995).
Hyatt, G.A., Schmitt, E.A., Fadool, J.M. & Dowling, J.E. Retinoic acid alters photoreceptor development in vivo. Proc. Natl. Acad. Sci. USA 93, 13298–13303 (1996).
Acknowledgements
We are grateful for funding from the UK Medical Research Council (COEN grant MR/J006904/1 to D.C.R. and C.J.O.'K.), the Wellcome Trust (Principal Fellowship 095317/Z/11/Z, to D.C.R., and strategic award 100140), the National Institute of Health Research Biomedical Research Unit in Dementia at Addenbrooke's Hospital, the TAMAHUD project (European Community FP6 grant no. 03472 under the Thematic Call LSH-2005-2.1.3-8 “Early markers and new targets for neurodegenerative diseases”) and the NEUROMICS project (European Community Seventh Framework Programme under grant agreement no. 2012-305121). We thank J.L. Marsh, N. Perrimon and the Vienna Drosophila RNAi Center for fly stocks; M. Renna and S. Luo for helpful comments; M. Lichtenberg for help with flow cytometry assays; F. Siddiqi and M. Garcia-Arencibia for help with primary cultures; W. Fecke for advice and assistance; and S. Gotta for help in high-resolution MS analysis of compounds.
Author information
Authors and Affiliations
Contributions
M.J.-S. performed most post-screen cell biology experiments. W.L. and S.I. performed the Drosophila experiments. M.H. and B.S. performed the cell-based screen. A.F., T.E.D. and C.X performed the zebrafish experiments, and A.F. supervised these. A.T., E.G.-C., V.P.S. and R.S. performed the bioinformatics analyses. F.M. performed the chaperone transcription array experiments and nonradioactive pulse-chase. E.G.-C. and F.H. participated in experimental design of the screen. F.H., G.L., D.D., L.M. and G.P. generated and validated the stable cell lines for the screen. G.M., C.C. and A.N. synthesized and analyzed the compounds. M.A. performed the selection of compound for high-throughput screening and supported the hit-to-lead optimization by in silico drug design methodologies. V.P. optimized glutaminyl cyclase enzymatic assays for compound screening. G.L.S. and N.P.C. performed in vitro ADME experiments. C.S. provided support for experiments at Siena Biotech. C.O.K. supervised Drosophila experiments. G.P. also supervised molecular biology activities at Siena Biotech. A.C. supervised primary screen and chemical biology. D.C.R. supervised cell biology, Drosophila and zebrafish experiments. D.C.R and A.C. conceived the project and coordinated work between sites with assistance from G.P., M.J.-S. and D.C.R., and A.C drafted the manuscript, which was commented on by all authors.
Corresponding authors
Ethics declarations
Competing interests
A.T., E.G.-C., G.L., F.H., D.D., L.M., G.P., G.M., C.C., A.N., M.A., G.L.S., N.P.C., V.P., C.S. and A.C. were employed by Siena Biotech; M.H. and B.S. were employed by Cenix BioScience GmbH.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Figures 1–18 and Supplementary Table 1 (PDF 4464 kb)
Supplementary Dataset 1
List of 257 human genes obtained in HEK293 siRNA screen and its validation in Drosophila (PDF 240 kb)
Supplementary Dataset 2
List of RNAi Drosophila lines that rescued Q48-eyedegeneration and their effect on GFP levels (PDF 81 kb)
Supplementary Dataset 3
Complete results from heat shock protein PCR array (PDF 116 kb)
Supplementary Note 1
Details for primary high-throughput siRNA screen development and analysis (PDF 72 kb)
Supplementary Note 2
Detailed synthesis and characterization of QPCT inhibitors (PDF 147 kb)
Rights and permissions
About this article
Cite this article
Jimenez-Sanchez, M., Lam, W., Hannus, M. et al. siRNA screen identifies QPCT as a druggable target for Huntington's disease. Nat Chem Biol 11, 347–354 (2015). https://doi.org/10.1038/nchembio.1790
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.1790
This article is cited by
-
Modifier pathways in polyglutamine (PolyQ) diseases: from genetic screens to drug targets
Cellular and Molecular Life Sciences (2022)
-
Genome-wide association studies of metabolites in Finnish men identify disease-relevant loci
Nature Communications (2022)
-
α-Catenin levels determine direction of YAP/TAZ response to autophagy perturbation
Nature Communications (2021)
-
Felodipine induces autophagy in mouse brains with pharmacokinetics amenable to repurposing
Nature Communications (2019)
-
Glutaminyl cyclase is an enzymatic modifier of the CD47- SIRPα axis and a target for cancer immunotherapy
Nature Medicine (2019)