Abstract
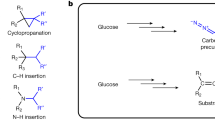
The terminal alkyne is a functionality widely used in organic synthesis, pharmaceutical science, material science and bioorthogonal chemistry. This functionality is also found in acetylenic natural products, but the underlying biosynthetic pathways for its formation are not well understood. Here we report the characterization of what is to our knowledge the first carrier protein–dependent terminal alkyne biosynthetic machinery in microbes. We further demonstrate that this enzymatic machinery can be exploited for the in situ generation and incorporation of terminal alkynes into two natural product scaffolds in Escherichia coli. These results highlight the prospect for tagging major classes of natural products, including polyketides and polyketide/nonribosomal peptide hybrids, using biosynthetic pathway engineering.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Bindman, N.A. & van der Donk, W.A. A general method for fluorescent labeling of the N-termini of lanthipeptides and its application to visualize their cellular localization. J. Am. Chem. Soc. 135, 10362–10371 (2013).
Sin, N., Meng, L., Auth, H. & Crews, C.M. Eponemycin analogues: syntheses and use as probes of angiogenesis. Bioorg. Med. Chem. 6, 1209–1217 (1998).
Harvey, C.J., Puglisi, J.D., Pande, V.S., Cane, D.E. & Khosla, C. Precursor directed biosynthesis of an orthogonally functional erythromycin analogue: selectivity in the ribosome macrolide binding pocket. J. Am. Chem. Soc. 134, 12259–12265 (2012).
Ross, C., Scherlach, K., Kloss, F. & Hertweck, C. The molecular basis of conjugated polyyne biosynthesis in phytopathogenic bacteria. Angew. Chem. Int. Edn Engl. 53, 7794–7798 (2014).
Thirumurugan, P., Matosiuk, D. & Jozwiak, K. Click chemistry for drug development and diverse chemical-biology applications. Chem. Rev. 113, 4905–4979 (2013).
Sandy, M., Zhu, X., Rui, Z. & Zhang, W. Characterization of AntB, a promiscuous acyltransferase involved in antimycin biosynthesis. Org. Lett. 15, 3396–3399 (2013).
Dupuis, S.N. et al. Synthetic diversification of natural products: semi-synthesis and evaluation of triazole jadomycins. Chem. Sci. 3, 1640–1644 (2012).
Yan, Y. et al. Multiplexing of combinatorial chemistry in antimycin biosynthesis: expansion of molecular diversity and utility. Angew. Chem. Int. Edn Engl. 52, 12308–12312 (2013).
Martín, J.F., Casqueiro, J. & Liras, P. Secretion systems for secondary metabolites: how producer cells send out messages of intercellular communication. Curr. Opin. Microbiol. 8, 282–293 (2005).
Minto, R.E. & Blacklock, B.J. Biosynthesis and function of polyacetylenes and allied natural products. Prog. Lipid Res. 47, 233–306 (2008).
Yamaguchi, M., Park, H.J., Ishizuka, S., Omata, K. & Hirama, M. Chemistry and antimicrobial activity of caryoynencins analogs. J. Med. Chem. 38, 5015–5022 (1995).
Shanklin, J., Guy, J.E., Mishra, G. & Lindqvist, Y. Desaturases: emerging models for understanding functional diversification of diiron-containing enzymes. J. Biol. Chem. 284, 18559–18563 (2009).
Gagné, S.J., Reed, D.W., Gray, G.R. & Covello, P.S. Structural control of chemoselectivity, stereoselectivity, and substrate specificity in membrane-bound fatty acid acetylenases and desaturases. Biochemistry 48, 12298–12304 (2009).
Haritos, V.S. et al. The convergent evolution of defensive polyacetylenic fatty acid biosynthesis genes in soldier beetles. Nat. Commun. 3, 1150 (2012).
Blacklock, B.J., Scheffler, B.E., Shepard, M.R., Jayasuriya, N. & Minto, R.E. Functional diversity in fungal fatty acid synthesis: the first acetylenase from the Pacific golden chanterelle, Cantharellus formosus. J. Biol. Chem. 285, 28442–28449 (2010).
Fritsche, K. et al. Biosynthetic genes and activity spectrum of antifungal polyynes from Collimonas fungivorans Ter331. Environ. Microbiol. 16, 1334–1345 (2014).
Lee, M. et al. Identification of non-heme diiron proteins that catalyze triple bond and epoxy group formation. Science 280, 915–918 (1998).
Edwards, D.J. et al. Structure and biosynthesis of the jamaicamides, new mixed polyketide-peptide neurotoxins from the marine cyanobacterium Lyngbya majuscula. Chem. Biol. 11, 817–833 (2004).
Jones, A.C. et al. Genomic insights into the physiology and ecology of the marine filamentous cyanobacterium Lyngbya majuscula. Proc. Natl. Acad. Sci. USA 108, 8815–8820 (2011).
Dorrestein, P.C. et al. Activity screening of carrier domains within nonribosomal peptide synthetases using complex substrate mixtures and large molecule mass spectrometry. Biochemistry 45, 1537–1546 (2006).
Shanklin, J. & Cahoon, E.B. Desaturation and related modifications of fatty acids. Annu. Rev. Plant Physiol. 49, 611–641 (1998).
Zhang, W., Bolla, M.L., Kahne, D. & Walsh, C.T. A three enzyme pathway for 2-amino-3-hydroxycyclopent-2-enone formation and incorporation in natural product biosynthesis. J. Am. Chem. Soc. 132, 6402–6411 (2010).
Wada, H., Avelange-Macherel, M.H. & Murata, N. The desA gene of the cyanobacterium Synechocystis sp. strain PCC6803 is the structural gene for Δ12 desaturase. J. Bacteriol. 175, 6056–6058 (1993).
Nakamura, M.T. & Nara, T.Y. Structure, function, and dietary regulation of Δ6, Δ5, and Δ9 desaturases. Annu. Rev. Nutr. 24, 345–376 (2004).
Wada, H., Schmidt, H., Heinz, E. & Murata, N. In vitro ferredoxin-dependent desaturation of fatty acids in cyanobacterial thylakoid membranes. J. Bacteriol. 175, 544–547 (1993).
Thompson, G.A. et al. Primary structures of the precursor and mature forms of stearoyl-acyl carrier protein desaturase from safflower embryos and requirement of ferredoxin for enzyme activity. Proc. Natl. Acad. Sci. USA 88, 2578–2582 (1991).
Shanklin, J. & Somerville, C. Stearoyl-acyl-carrier-protein desaturase from higher plants is structurally unrelated to the animal and fungal homologs. Proc. Natl. Acad. Sci. USA 88, 2510–2514 (1991).
Yin, J., Lin, A.J., Golan, D.E. & Walsh, C.T. Site-specific protein labeling by Sfp phosphopantetheinyl transferase. Nat. Protoc. 1, 280–285 (2006).
Morita, H. et al. Synthesis of unnatural alkaloid scaffolds by exploiting plant polyketide synthase. Proc. Natl. Acad. Sci. USA 108, 13504–13509 (2011).
Pfeifer, B.A., Admiraal, S.J., Gramajo, H., Cane, D.E. & Khosla, C. Biosynthesis of complex polyketides in a metabolically engineered strain of E. coli. Science 291, 1790–1792 (2001).
Zhang, W., Li, Y. & Tang, Y. Engineered biosynthesis of bacterial aromatic polyketides in Escherichia coli. Proc. Natl. Acad. Sci. USA 105, 20683–20688 (2008).
Shimo, T., Matsuzaki, S. & Somekawa, K. Intramolecular photoreactions of 6-(ω-alkenyl)-4-methoxy-2-pyrones. J. Heterocycl. Chem. 31, 387–390 (1994).
Wilson, M.C. & Moore, B.S. Beyond ethylmalonyl-CoA: the functional role of crotonyl-CoA carboxylase/reductase homologs in expanding polyketide diversity. Nat. Prod. Rep. 29, 72–86 (2012).
Sandy, M., Rui, Z., Gallagher, J. & Zhang, W. Enzymatic synthesis of dilactone scaffold of antimycins. ACS Chem. Biol. 7, 1956–1961 (2012).
Pyke, K.A. Plastid division and development. Plant Cell 11, 549–556 (1999).
Shanklin, J., Whittle, E. & Fox, B.G. Eight histidine residues are catalytically essential in a membrane-associated iron enzyme, stearoyl-CoA desaturase, and are conserved in alkane hydroxylase and xylene monooxygenase. Biochemistry 33, 12787–12794 (1994).
Shieh, P., Hangauer, M.J. & Bertozzi, C.R. Fluorogenic azidofluoresceins for biological imaging. J. Am. Chem. Soc. 134, 17428–17431 (2012).
Shieh, P., Siegrist, M.S., Cullen, A.J. & Bertozzi, C.R. Imaging bacterial peptidoglycan with near-infrared fluorogenic azide probes. Proc. Natl. Acad. Sci. USA 111, 5456–5461 (2014).
Grammel, M. & Hang, H.C. Chemical reporters for biological discovery. Nat. Chem. Biol. 9, 475–484 (2013).
Neef, A.B. & Luedtke, N.W. Dynamic metabolic labeling of DNA in vivo with arabinosyl nucleosides. Proc. Natl. Acad. Sci. USA 108, 20404–20409 (2011).
Torella, J.P. et al. Tailored fatty acid synthesis via dynamic control of fatty acid elongation. Proc. Natl. Acad. Sci. USA 110, 11290–11295 (2013).
Liu, J., Zhu, X., Seipke, R.F. & Zhang, W. Biosynthesis of antimycins with a reconstituted 3-formamidosalicylate pharmacophore in Escherichia coli. ACS Synth. Biol. 10.1021/sb5003136 (2014).
Acknowledgements
This research was financially supported by the Pew Scholars Program and University of California Cancer Research Coordinating Committee funds. We thank I. Abe (University of Tokyo) for providing hspks1, S. Bauer (University of California–Berkeley) for assisting with LC/HRMS analysis, J. Pelton (University of California–Berkeley) for helping with NMR spectroscopic analysis, J. Chung (University of California–Berkeley) for helping with compound purification and L. Zhang (University of California–Berkeley) for providing scACP.
Author information
Authors and Affiliations
Contributions
X.Z. designed the experiments, performed all experiments, analyzed the data and wrote the manuscript. J.L. constructed plasmids for antimycin production in E. coli. W.Z. designed the experiments, analyzed the data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Tables 1–4 and Supplementary Figures 1–23. (PDF 4454 kb)
Rights and permissions
About this article
Cite this article
Zhu, X., Liu, J. & Zhang, W. De novo biosynthesis of terminal alkyne-labeled natural products. Nat Chem Biol 11, 115–120 (2015). https://doi.org/10.1038/nchembio.1718
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.1718
This article is cited by
-
Mining genomes to illuminate the specialized chemistry of life
Nature Reviews Genetics (2021)
-
Bioorthogonal chemistry
Nature Reviews Methods Primers (2021)
-
Next-generation physiology approaches to study microbiome function at single cell level
Nature Reviews Microbiology (2020)
-
Recent Advances in Re-engineering Modular PKS and NRPS Assembly Lines
Biotechnology and Bioprocess Engineering (2020)
-
Discovery of a pathway for terminal-alkyne amino acid biosynthesis
Nature (2019)