Abstract
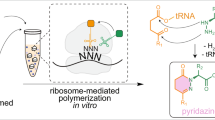
Peptide antibiotics are typically biosynthesized by one of two distinct machineries in a ribosome-dependent or ribosome-independent manner. Pheganomycin (PGM (1)) and related analogs consist of the nonproteinogenic amino acid (S)-2-(3,5-dihydroxy-4-hydroxymethyl)phenyl-2-guanidinoacetic acid (2) and a proteinogenic core peptide, making their origin uncertain. We report the identification of the biosynthetic gene cluster from Streptomyces cirratus responsible for PGM production. Unexpectedly, the cluster contains a gene encoding multiple precursor peptides along with several genes plausibly encoding enzymes for the synthesis of amino acid 2. We identified PGM1, which has an ATP-grasp domain, as potentially capable of linking the precursor peptides with 2, and validate this hypothesis using deletion mutants and in vitro reconstitution. We document PGM1's substrate permissivity, which could be rationalized by a large binding pocket as confirmed via structural and mutagenesis experiments. This is to our knowledge the first example of cooperative peptide synthesis achieved by ribosomes and peptide ligases using a peptide nucleophile.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Humphries, R.M., Pollett, S. & Sakoulas, G. A current perspective on daptomycin for the clinical microbiologist. Clin. Microbiol. Rev. 26, 759–780 (2013).
Arnison, P.G. et al. Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 30, 108–160 (2013).
Koglin, A. & Walsh, C.T. Structural insights into nonribosomal peptide enzymatic assembly lines. Nat. Prod. Rep. 26, 987–1000 (2009).
Giessen, T.W. & Marahiel, M.A. Ribosome-independent biosynthesis of biologically active peptides: application of synthetic biology to generate structural diversity. FEBS Lett. 586, 2065–2075 (2012).
Maruyama, C. et al. A stand-alone adenylation domain forms amide bonds in streptothricin biosynthesis. Nat. Chem. Biol. 8, 791–797 (2012).
Kadi, N., Oves-Costales, D., Barona-Gomez, F. & Challis, G.L. A new family of ATP-dependent oligomerization-macrocyclization biocatalysts. Nat. Chem. Biol. 3, 652–656 (2007).
Suzukake-Tsuchiya, K., Hori, M., Shimada, N. & Hamada, M. Mode of action of deoxypheganomycin D on Mycobacterium smegmatis ATCC 607. J. Antibiot. (Tokyo) 41, 675–683 (1988).
Crone, W.J.K., Leeper, F.J. & Truman, A.W. Identification and characterisation of the gene cluster for the anti-MRSA antibiotic bottromycin: expanding the biosynthetic diversity of ribosomal peptides. Chemical Science 3, 3516–3521 (2012).
Huo, L., Rachid, S., Stadler, M., Wenzel, S.C. & Muller, R. Synthetic biotechnology to study and engineer ribosomal bottromycin biosynthesis. Chem. Biol. 19, 1278–1287 (2012).
Hubbard, B.K. & Walsh, C.T. Vancomycin assembly: nature's way. Angew. Chem. Int. Edn Engl. 42, 730–765 (2003).
Donia, M.S., Ravel, J. & Schmidt, E.W. A global assembly line for cyanobactins. Nat. Chem. Biol. 4, 341–343 (2008).
Chen, H., Tseng, C.C., Hubbard, B.K. & Walsh, C.T. Glycopeptide antibiotic biosynthesis: enzymatic assembly of the dedicated amino acid monomer (S)-3,5-dihydroxyphenylglycine. Proc. Natl. Acad. Sci. USA 98, 14901–14906 (2001).
Anzai, H. et al. Replacement of Streptomyces hygroscopicus genomic segments with in vitro altered DNA sequences. J. Antibiot. (Tokyo) 41, 226–233 (1988).
Fawaz, M.V., Topper, M.E. & Firestine, S.M. The ATP-grasp enzymes. Bioorg. Chem. 39, 185–191 (2011).
Yamaguchi, H. et al. Three-dimensional structure of the glutathione synthetase from Escherichia coli B at 2.0 Å resolution. J. Mol. Biol. 229, 1083–1100 (1993).
Fan, C., Moews, P.C., Shi, Y., Walsh, C.T. & Knox, J.R. A common fold for peptide synthetases cleaving ATP to ADP: glutathione synthetase and D-alanine:D-alanine ligase of Escherichia coli. Proc. Natl. Acad. Sci. USA 92, 1172–1176 (1995).
Tabata, K., Ikeda, H. & Hashimoto, S. ywfE in Bacillus subtilis codes for a novel enzyme, L-amino acid ligase. J. Bacteriol. 187, 5195–5202 (2005).
Kino, K., Arai, T. & Tateiwa, D. A novel L-amino acid ligase from Bacillus subtilis NBRC3134 catalyzed oligopeptide synthesis. Biosci. Biotechnol. Biochem. 74, 129–134 (2010).
Taguchi, S., Mita, K., Ichinohe, K. & Hashimoto, S. Targeted engineering of the antibacterial peptide apidaecin, based on an in vivo monitoring assay system. Appl. Environ. Microbiol. 75, 1460–1464 (2009).
Holm, L. & Sander, C. Dali: a network tool for protein structure comparison. Trends Biochem. Sci. 20, 478–480 (1995).
Sampei, G. et al. Crystal structure of glycinamide ribonucleotide synthetase, PurD, from thermophilic eubacteria. J. Biochem. 148, 429–438 (2010).
Thoden, J.B., Holden, H.M., Paritala, H. & Firestine, S.M. Structural and functional studies of Aspergillus clavatus N5-carboxyaminoimidazole ribonucleotide synthetase. Biochemistry 49, 752–760 (2010).
Thoden, J.B., Firestine, S.M., Benkovic, S.J. & Holden, H.M. PurT-encoded glycinamide ribonucleotide transformylase. J. Biol. Chem. 277, 23898–23908 (2002).
Suzuki, M. et al. The structure of L-amino-acid ligase from Bacillus licheniformis. Acta Crystallogr. D Biol. Crystallogr. 68, 1535–1540 (2012).
Wang, W., Kappock, T.J., Stubbe, J. & Ealick, S.E. X-ray crystal structure of glycinamide ribonucleotide synthetase from Escherichia coli. Biochemistry 37, 15647–15662 (1998).
Bateman, A. et al. The Pfam protein families database. Nucleic Acids Res. 28, 263–266 (2000).
Teixeira, L.K. & Reed, S.I. Ubiquitin ligases and cell cycle control. Annu. Rev. Biochem. 82, 387–414 (2013).
Mazmanian, S.K., Liu, G., Ton-That, H. & Schneewind, O. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science 285, 760–763 (1999).
Kane, P.M. et al. Protein splicing converts the yeast TFP1 gene product to the 69-kD subunit of the vacuolar H+-adenosine triphosphatase. Science 250, 651–657 (1990).
Chan, A.O. et al. Modification of N-terminal α-amino groups of peptides and proteins using ketenes. J. Am. Chem. Soc. 134, 2589–2598 (2012).
Ong, S.E. & Mann, M. Mass spectrometry-based proteomics turns quantitative. Nat. Chem. Biol. 1, 252–262 (2005).
Wagner, A.M. et al. N-terminal protein modification using simple aminoacyl transferase substrates. J. Am. Chem. Soc. 133, 15139–15147 (2011).
Pan, Y. et al. N-terminal labeling of peptides by trypsin-catalyzed ligation for quantitative proteomics. Angew. Chem. Int. Edn Engl. 52, 9205–9209 (2013).
Jursic, B.S., Neumann, D. & McPherson, A. Preparation of N-formamidinylamino acids from amino and formamidinesulfinic acids. Synthesis 32, 1656–1658 (2000).
Zerbino, D.R. & Birney, E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18, 821–829 (2000).
Noguchi, H., Taniguchi, T. & Itoh, T. MetaGeneAnnotator: detecting species-specific patterns of ribosomal binding site for precise gene prediction in anonymous prokaryotic and phage genomes. DNA Res. 15, 387–396 (2008).
Vara, J., Lewandowska-Skarbek, M., Wang, Y.G., Donadio, S. & Hutchinson, C.R. Cloning of genes governing the deoxysugar portion of the erythromycin biosynthesis pathway in Saccharopolyspora erythraea (Streptomyces erythreus). J. Bacteriol. 171, 5872–5881 (1989).
Hopwood, D.A. et al. Gene manipulation of Streptomyces: a Laboratory Manual (Cold Spring Harbor Laboratory Press, New York, 1985).
Rice, L.M., Earnest, T.N. & Brunger, A.T. Single-wavelength anomalous diffraction phasing revisited. Acta Crystallogr. D Biol. Crystallogr. 56, 1413–1420 (2000).
Kabsch, W. XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010).
Adams, P.D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Afonine, P.V. et al. Towars automated crystallographic structure refinement with phenix. refine. Acta Crystallogr. D Biol. Crystallogr. 68, 352–367 (2012).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Chen, V.B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010).
Brunger, A.T. Version 1.2 of the crystallography and NMR system. Nat. Protoc. 2, 2728–2733 (2007).
Trott, O. & Olson, A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J. Comput. Chem. 31, 455–461 (2010).
Acknowledgements
This study was supported by a Grant-in-Aid for Scientific Research (23108101 and 25560397 to T.D. and 25108710 to H.M.) from the Japan Society for the Promotion of Science. We thank M. Igarashi from the Institute of Microbial Chemistry and S. Taguchi from Hokkaido University for providing us with S. cirratus and apidaecin, respectively.
Author information
Authors and Affiliations
Contributions
M.N., H.M. and T.D. designed the research. M.N., T.M. and K.O. performed in vitro experiments. T.M., S.O. and H.M. carried out crystallography. I.S. and H.I. synthesized substrates and performed NMR studies. M.N., Y.H., C.M. and Y.S. analyzed products by LC/MS. J.I. and T.D. identified the gene cluster. M.N., H.M. and T.D. analyzed data and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Tables 1–8 and Supplementary Figures 1–39. (PDF 4443 kb)
Rights and permissions
About this article
Cite this article
Noike, M., Matsui, T., Ooya, K. et al. A peptide ligase and the ribosome cooperate to synthesize the peptide pheganomycin. Nat Chem Biol 11, 71–76 (2015). https://doi.org/10.1038/nchembio.1697
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.1697
This article is cited by
-
Phylogenomic analysis of the diversity of graspetides and proteins involved in their biosynthesis
Biology Direct (2022)
-
Acyltransferase that catalyses the condensation of polyketide and peptide moieties of goadvionin hybrid lipopeptides
Nature Chemistry (2020)
-
Plant-like biosynthesis of isoquinoline alkaloids in Aspergillus fumigatus
Nature Chemical Biology (2016)
-
Structure and activity relationships of the anti-Mycobacterium antibiotics resorcinomycin and pheganomycin
The Journal of Antibiotics (2016)