Abstract
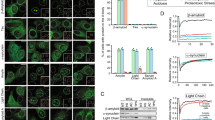
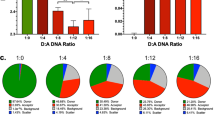
We previously discovered a small-molecule inducer of cell death, named 1541, that noncovalently self-assembles into chemical fibrils ('chemi-fibrils') and activates procaspase-3 in vitro. We report here that 1541-induced cell death is caused by the fibrillar rather than the soluble form of the drug. A short hairpin RNA screen reveals that knockdown of genes involved in endocytosis, vesicle trafficking and lysosomal acidification causes partial 1541 resistance. We confirm the role of these pathways using pharmacological inhibitors. Microscopy shows that the fluorescent chemi-fibrils accumulate in punctae inside cells that partially colocalize with lysosomes. Notably, the chemi-fibrils bind and induce liposome leakage in vitro, suggesting they may do the same in cells. The chemi-fibrils induce extensive proteolysis including caspase substrates, yet modulatory profiling reveals that chemi-fibrils form a distinct class from existing inducers of cell death. The chemi-fibrils share similarities with proteinaceous fibrils and may provide insight into their mechanism of cellular toxicity.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Jucker, M. & Walker, L.C. Self-propagation of pathogenic protein aggregates in neurodegenerative diseases. Nature 501, 45–51 (2013).
Eisenberg, D. & Jucker, M. The amyloid state of proteins in human diseases. Cell 148, 1188–1203 (2012).
Wolan, D.W., Zorn, J.A., Gray, D.C. & Wells, J.A. Small-molecule activators of a proenzyme. Science 326, 853–858 (2009).
Zorn, J.A., Wille, H., Wolan, D.W. & Wells, J.A. Self-assembling small molecules form nanofibrils that bind procaspase-3 to promote activation. J. Am. Chem. Soc. 133, 19630–19633 (2011).
Zorn, J.A., Wolan, D.W., Agard, N.J. & Wells, J.A. Fibrils colocalize caspase-3 with procaspase-3 to foster maturation. J. Biol. Chem. 287, 33781–33795 (2012).
Bassik, M.C. et al. Rapid creation and quantitative monitoring of high coverage shRNA libraries. Nat. Methods 6, 443–445 (2009).
Bassik, M.C. et al. A systematic Mammalian genetic interaction map reveals pathways underlying ricin susceptibility. Cell 152, 909–922 (2013).
Kampmann, M., Bassik, M.C. & Weissman, J.S. Integrated platform for genome-wide screening and construction of high-density genetic interaction maps in mammalian cells. Proc. Natl. Acad. Sci. USA 110, E2317–E2326 (2013).
Weiss, W.A., Taylor, S.S. & Shokat, K.M. Recognizing and exploiting differences between RNAi and small-molecule inhibitors. Nat. Chem. Biol. 3, 739–744 (2007).
Crawford, E.D. et al. The DegraBase: a database of proteolysis in healthy and apoptotic human cells. Mol. Cell. Proteomics 12, 813–824 (2013).
Mahrus, S. et al. Global sequencing of proteolytic cleavage sites in apoptosis by specific labeling of protein N termini. Cell 134, 866–876 (2008).
Wiita, A.P., Seaman, J.E. & Wells, J.A. Global analysis of cellular proteolysis by selective enzymatic labeling of protein N-termini. Methods Enzymol. 544, 327–358 (2014).
Wolpaw, A.J. et al. Modulatory profiling identifies mechanisms of small molecule–induced cell death. Proc. Natl. Acad. Sci. USA 108, E771–E780 (2011).
Holmes, B.B. et al. Heparan sulfate proteoglycans mediate internalization and propagation of specific proteopathic seeds. Proc. Natl. Acad. Sci. USA 110, E3138–E3147 (2013).
Owen, S.C., Doak, A.K., Wassam, P., Shoichet, M.S. & Shoichet, B.K. Colloidal aggregation affects the efficacy of anticancer drugs in cell culture. ACS Chem. Biol. 7, 1429–1435 (2012).
Owen, S.C. et al. Colloidal drug formulations can explain “bell-shaped” concentration-response curves. ACS Chem. Biol. 9, 777–784 (2014).
Kampmann, M., Bassik, M.C. & Weissman, J.S. Functional genomics platform for pooled screening and generation of mammalian genetic interaction maps. Nat. Protoc. 9, 1825–1847 (2014).
Tisdale, E.J., Bourne, J.R., Khosravi-Far, R., Der, C.J. & Balch, W.E. GTP-binding mutants of rab1 and rab2 are potent inhibitors of vesicular transport from the endoplasmic reticulum to the Golgi complex. J. Cell Biol. 119, 749–761 (1992).
Bucci, C., Thomsen, P., Nicoziani, P., McCarthy, J. & van Deurs, B. Rab7: a key to lysosome biogenesis. Mol. Biol. Cell 11, 467–480 (2000).
Stenmark, H., Vitale, G., Ullrich, O. & Zerial, M. Rabaptin-5 is a direct effector of the small GTPase Rab5 in endocytic membrane fusion. Cell 83, 423–432 (1995).
Seto, S., Tsujimura, K. & Koide, Y. Rab GTPases regulating phagosome maturation are differentially recruited to mycobacterial phagosomes. Traffic 12, 407–420 (2011).
Stenmark, H. Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 10, 513–525 (2009).
Münch, C., O'Brien, J. & Bertolotti, A. Prion-like propagation of mutant superoxide dismutase-1 misfolding in neuronal cells. Proc. Natl. Acad. Sci. USA 108, 3548–3553 (2011).
Weinstein, J.N., Yoshikami, S., Henkart, P., Blumenthal, R. & Hagins, W.A. Liposome-cell interaction: transfer and intracellular release of a trapped fluorescent marker. Science 195, 489–492 (1977).
Shimbo, K. et al. Quantitative profiling of caspase-cleaved substrates reveals different drug-induced and cell-type patterns in apoptosis. Proc. Natl. Acad. Sci. USA 109, 12432–12437 (2012).
Wiita, A.P. et al. Global cellular response to chemotherapy-induced apoptosis. Elife 2, e01236 (2013).
Fraley, C. & Raftery, A.E. Model-based clustering, discriminant analysis, and density estimation. J. Am. Stat. Assoc. 97, 611–631 (2002).
Duewell, P. et al. NLRP3 inflammasomes are required for atherogenesis and activated by cholesterol crystals. Nature 464, 1357–1361 (2010).
Halle, A. et al. The NALP3 inflammasome is involved in the innate immune response to amyloid-β. Nat. Immunol. 9, 857–865 (2008).
Huff, M.E., Balch, W.E. & Kelly, J.W. Pathological and functional amyloid formation orchestrated by the secretory pathway. Curr. Opin. Struct. Biol. 13, 674–682 (2003).
Novitskaya, V., Bocharova, O.V., Bronstein, I. & Baskakov, I.V. Amyloid fibrils of mammalian prion protein are highly toxic to cultured cells and primary neurons. J. Biol. Chem. 281, 13828–13836 (2006).
Yao, J. et al. Neuroprotection by cyclodextrin in cell and mouse models of Alzheimer disease. J. Exp. Med. 209, 2501–2513 (2012).
Walsh, D.M. & Selkoe, D.J. Aβ oligomers—a decade of discovery. J. Neurochem. 101, 1172–1184 (2007).
Acknowledgements
We would like to thank F. Brodsky, B. Shoichet, W. Degrado, M. Zhuang, A. Wiita, Z. Hill, J.T. Koerber, N. Thomsen, J. Watts, S.-A. Mok and J. Rettenmaier for insightful discussions and/or critical reading of the manuscript. A special thanks to Y. Chen (cell culture and laboratory practices expertise), Y. Cheng and M. Braunfield (EM), A. Doak (DLS), H. Tran (yeast expertise), D. Larsen (live cell imaging), J. Lund (deep sequencing), M. Hornsby and K. Verba (fluorescence) and T. Matsuguchi (qPCR) for technical help. This work was supported, in whole or in part, by US National Institutes of Health grant R01 CA136779 (to J.A.W.), R01 CA097061 (to B.R.S.) and F32AI095062 (to V.J.V.) and by the Howard Hughes Medical Institute (to B.R.S. and J.S.W.). J.A.Z. received an Achievement Rewards for College Scientists Foundation Award and a Schleroderma Research Foundation Evnin-Wright Fellowship. M.K. was supported by a postdoctoral fellowship from the Jane Coffin Childs Memorial Fund. O.J. is the recipient of a Banting Postdoctoral Fellowship funded by the Canadian Institutes of Health Research and the Government of Canada. O.J. and M.K. both received a fellowship from the University of California–San Francisco Program for Breakthrough Biomedical Research, which is funded in part by the Sandler Foundation.
Author information
Authors and Affiliations
Contributions
O.J. performed the cell culture experiments, EM, DLS, flow cytometry and fluorescence microscopy under J.A.W.'s supervision. O.J. performed the shRNA screen and made the stable cell lines, with help and guidance from M.K. and M.C.B. and supervision of J.S.W.; M.K. analyzed the deep-sequencing data. J.A.Z. synthesized the 1541 analogs and provided general expertise on the project. A.L.R. performed the crystallography and structure determination. V.J.V. performed the liposome leakage assays. K. Shimbo and N.J.A. performed the degradomics experiments, and O.J. compiled the results. K. Shimada performed the modulatory profiling experiments under B.R.S.'s supervision. O.J. and J.A.W. wrote the manuscript with contributions from M.K., J.A.Z. and B.R.S., with input from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results and Supplementary Figures 1–22. (PDF 12319 kb)
Supplementary Data Set 1
P values for all genes tested in the shRNA screen. (XLSX 324 kb)
Supplementary Data Set 2
Identification of proteolytic fragments generated during chemi-fibril induced cell death. (XLSX 3535 kb)
Supplementary Video 1
Live cell imaging of HeLa cells treated with chemi-fibrils. (MOV 5773 kb)
Supplementary Video 2
Live cell imaging of HeLa cells treated with chemi-fibrils. (MOV 13992 kb)
Rights and permissions
About this article
Cite this article
Julien, O., Kampmann, M., Bassik, M. et al. Unraveling the mechanism of cell death induced by chemical fibrils. Nat Chem Biol 10, 969–976 (2014). https://doi.org/10.1038/nchembio.1639
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.1639
This article is cited by
-
Non-apoptotic cell death induction via sapogenin based supramolecular particles
Scientific Reports (2022)
-
A new era for understanding amyloid structures and disease
Nature Reviews Molecular Cell Biology (2018)
-
Enzymatic formation of curcumin in vitro and in vivo
Nano Research (2018)
-
Allosteric sensitization of proapoptotic BAX
Nature Chemical Biology (2017)
-
Mitochondria localization induced self-assembly of peptide amphiphiles for cellular dysfunction
Nature Communications (2017)