Abstract
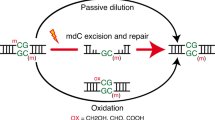
Ten eleven translocation (Tet) enzymes oxidize the epigenetically important DNA base 5-methylcytosine (mC) stepwise to 5-hydroxymethylcytosine (hmC), 5-formylcytosine and 5-carboxycytosine. It is currently unknown whether Tet-induced oxidation is limited to cytosine-derived nucleobases or whether other nucleobases are oxidized as well. We synthesized isotopologs of all major oxidized pyrimidine and purine bases and performed quantitative MS to show that Tet-induced oxidation is not limited to mC but that thymine is also a substrate that gives 5-hydroxymethyluracil (hmU) in mouse embryonic stem cells (mESCs). Using MS-based isotope tracing, we show that deamination of hmC does not contribute to the steady-state levels of hmU in mESCs. Protein pull-down experiments in combination with peptide tracing identifies hmU as a base that influences binding of chromatin remodeling proteins and transcription factors, suggesting that hmU has a specific function in stem cells besides triggering DNA repair.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Smith, Z.D. & Meissner, A. DNA methylation: roles in mammalian development. Nat. Rev. Genet. 14, 204–220 (2013).
Franchini, D.M., Schmitz, K.M. & Petersen-Mahrt, S.K. 5-Methylcytosine DNA demethylation: more than losing a methyl group. Annu. Rev. Genet. 46, 419–441 (2012).
Tahiliani, M. et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324, 930–935 (2009).
Pfaffeneder, T. et al. The discovery of 5-formylcytosine in embryonic stem cell DNA. Angew. Chem. Int. Ed. Engl. 50, 7008–7012 (2011).
Ito, S. et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333, 1300–1303 (2011).
He, Y.F. et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333, 1303–1307 (2011).
Tan, L. & Shi, Y.G. Tet family proteins and 5-hydroxymethylcytosine in development and disease. Development 139, 1895–1902 (2012).
Maiti, A. & Drohat, A.C. Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: potential implications for active demethylation of CpG sites. J. Biol. Chem. 286, 35334–35338 (2011).
Bjelland, S. et al. Oxidation of thymine to 5-formyluracil in DNA: mechanisms of formation, structural implications, and base excision by human cell free extracts. Biochemistry 34, 14758–14764 (1995).
Mouret, J.F., Polverelli, M., Sarrazini, F. & Cadet, J. Ionic and radical oxidations of DNA by hydrogen peroxide. Chem. Biol. Interact. 77, 187–201 (1991).
Guo, J.U., Su, Y., Zhong, C., Ming, G.L. & Song, H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell 145, 423–434 (2011).
Cortellino, S. et al. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell 146, 67–79 (2011).
Nabel, C.S. et al. AID/APOBEC deaminases disfavor modified cytosines implicated in DNA demethylation. Nat. Chem. Biol. 8, 751–758 (2012).
Rangam, G., Schmitz, K.M., Cobb, A.J. & Petersen-Mahrt, S.K. AID enzymatic activity is inversely proportional to the size of cytosine C5 orbital cloud. PLoS ONE 7, e43279 (2012).
Jacobs, A.L. & Schär, P. DNA glycosylases: in DNA repair and beyond. Chromosoma 121, 1–20 (2012).
Zhang, Q.M. et al. DNA glycosylase activities for thymine residues oxidized in the methyl group are functions of the hNEIL1 and hNTH1 enzymes in human cells. DNA Repair (Amst.) 4, 71–79 (2005).
Burrows, C.J. Surviving an oxygen atmosphere: DNA damage and repair. ACS Symp. Ser. Am. Chem. Soc. 2009, 147–156 (2009).
Taghizadeh, K. et al. Quantification of DNA damage products resulting from deamination, oxidation and reaction with products of lipid peroxidation by liquid chromatography isotope dilution tandem mass spectrometry. Nat. Protoc. 3, 1287–1298 (2008).
Globisch, D. et al. Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PLoS ONE 5, e15367 (2010).
Kriaucionis, S. & Heintz, N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 324, 929–930 (2009).
Münzel, M. et al. Quantification of the sixth DNA base hydroxymethylcytosine in the brain. Angew. Chem. Int. Ed. Engl. 49, 5375–5377 (2010).
Liu, S. et al. Quantitative assessment of Tet-induced oxidation products of 5-methylcytosine in cellular and tissue DNA. Nucleic Acids Res. 41, 6421–6429 (2013).
Ficz, G. et al. FGF signaling inhibition in ESCs drives rapid genome-wide demethylation to the epigenetic ground state of pluripotency. Cell Stem Cell 13, 351–359 (2013).
Habibi, E. et al. Whole-genome bisulfite sequencing of two distinct interconvertible DNA methylomes of mouse embryonic stem cells. Cell Stem Cell 13, 360–369 (2013).
Leitch, H.G. et al. Naive pluripotency is associated with global DNA hypomethylation. Nat. Struct. Mol. Biol. 20, 311–316 (2013).
Hayashi, K., Ohta, H., Kurimoto, K., Aramaki, S. & Saitou, M. Reconstitution of the mouse germ cell specification pathway in culture by pluripotent stem cells. Cell 146, 519–532 (2011).
Ying, Q.L., Stavridis, M., Griffiths, D., Li, M. & Smith, A. Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat. Biotechnol. 21, 183–186 (2003).
Iurlaro, M. et al. A screen for hydroxymethylcytosine and formylcytosine binding proteins suggests functions in transcription and chromatin regulation. Genome Biol. 14, R119 (2013).
Spruijt, C.G. et al. Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell 152, 1146–1159 (2013).
Burley, G.A. et al. Directed DNA metallization. J. Am. Chem. Soc. 128, 1398–1399 (2006).
Rostovtsev, V.V., Green, L.G., Fokin, V.V. & Sharpless, K.B. A stepwise huisgen cycloaddition process: copper(I)-catalyzed regioselective “ligation” of azides and terminal alkynes. Angew. Chem. Int. Ed. 41, 2596–2599 (2002).
Thompson, A. et al. Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal. Chem. 75, 1895–1904 (2003); erratum 75, 4942 (2003); erratum 78, 4235 (2006).
Gygi, S.P. et al. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat. Biotechnol. 17, 994–999 (1999).
Frauer, C. et al. Recognition of 5-hydroxymethylcytosine by the Uhrf1 SRA domain. PLoS ONE 6, e21306 (2011).
Pichler, G. et al. Cooperative DNA and histone binding by Uhrf2 links the two major repressive epigenetic pathways. J. Cell. Biochem. 112, 2585–2593 (2011).
Lewis, H.L., Muhleman, D.R. & Ward, J.F. Serologic assay of DNA base damage. I. 5-Hydroxymethyldeoxyuridine, a radiation product of thymidine. Radiat. Res. 75, 305–316 (1978).
Boorstein, R.J. et al. Definitive identification of mammalian 5-hydroxymethyluracil DNA N-glycosylase activity as SMUG1. J. Biol. Chem. 276, 41991–41997 (2001).
Kavli, B., Otterlei, M., Slupphaug, G. & Krokan, H.E. Uracil in DNA—general mutagen, but normal intermediate in acquired immunity. DNA Repair (Amst.) 6, 505–516 (2007).
Santos, F. et al. Active demethylation in mouse zygotes involves cytosine deamination and base excision repair. Epigenetics Chromatin 6, 39 (2013).
Silva, J. & Smith, A. Capturing pluripotency. Cell 132, 532–536 (2008).
Moore, S.P.G., Toomire, K.J. & Strauss, P.R. DNA modifications repaired by base excision repair are epigenetic. DNA Repair (Amst.) 12, 1152–1158 (2013).
Schiesser, S. et al. Deamination, oxidation, and C–C bond cleavage reactivity of 5-hydroxymethylcytosine, 5-formylcytosine, and 5-carboxycytosine. J. Am. Chem. Soc. 135, 14593–14599 (2013).
Gierlich, J. et al. Click chemistry as a reliable method for the high-density postsynthetic functionalization of alkyne-modified DNA. Org. Lett. 8, 3639–3642 (2006).
Cao, H. & Wang, Y. Collisionally activated dissociation of protonated 2′-deoxycytidine, 2′-deoxyuridine, and their oxidatively damaged derivatives. J. Am. Soc. Mass Spectrom. 17, 1335–1341 (2006).
Wang, J. et al. Quantification of oxidative DNA lesions in tissues of Long-Evans Cinnamon rats by capillary high-performance liquid chromatography-tandem mass spectrometry coupled with stable isotope-dilution method. Anal. Chem. 83, 2201–2209 (2011).
Chen, T., Ueda, Y., Dodge, J.E., Wang, Z. & Li, E. Establishment and maintenance of genomic methylation patterns in mouse embryonic stem cells by Dnmt3a and Dnmt3b. Mol. Cell. Biol. 23, 5594–5605 (2003).
Cortazar, D. et al. Embryonic lethal phenotype reveals a function of TDG in maintaining epigenetic stability. Nature 470, 419–423 (2011).
Nagy, A., Rossant, J., Nagy, R., Abramow-Newerly, W. & Roder, J.C. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc. Natl. Acad. Sci. USA 90, 8424–8428 (1993).
Li, E., Bestor, T.H. & Jaenisch, R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69, 915–926 (1992).
Montanez, E. et al. Kindlin-2 controls bidirectional signaling of integrins. Genes Dev. 22, 1325–1330 (2008).
Ying, Q.L. & Smith, A.G. Defined conditions for neural commitment and differentiation. Methods Enzymol. 365, 327–341 (2003).
Williams, K. et al. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature 473, 343–348 (2011).
Kittler, R., Heninger, A.K., Franke, K., Habermann, B. & Buchholz, F. Production of endoribonuclease-prepared short interfering RNAs for gene silencing in mammalian cells. Nat. Methods 2, 779–784 (2005).
Yu, M. et al. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell 149, 1368–1380 (2012).
Acknowledgements
We thank the Excellence Cluster Center for Integrated Protein Science Munich (CiPSM) and the collaborative research centers SFB749, SFB646 and SFB1032 as well as German Research Foundation (DFG) grant CA275/8-4, the Volkswagen foundation, NGFNplus (01GS0870) and the Netherlands Organization for Scientific Research (NWO-VIDI) for financial support. T.P. and S.S. thank the Fonds der Chemischen Industrie for predoctoral fellowships. C.B. thanks the Boehringer Ingelheim Fonds for a predoctoral fellowship. G.K. thanks the Japan Society for the Promotion of Science (JSPS) for a postdoctoral fellowship for research abroad. We thank M. Moser (Max Planck Institute for Biochemistry) for providing R1- and C57Bl6/129–derived mESCs, G. Höfner and K.T. Wanner for their initial help with MS as well as M. Wirsing, L. Belzner and P. Laube for providing bioinformatic tools for data processing.
Author information
Authors and Affiliations
Contributions
T.P. synthesized MS standards, performed the sample preparation and ultra high-performance LC/MS/MS method development, did LC/MS analysis, interpreted data and performed statistical analysis. F.S. and N.R. performed mESC differentiation and isotope tracing experiments. M.W. and C.B. performed the HEK-293T experiments, did LC/MS analysis and interpreted data. C.B. performed qPCR and analyzed the data. S.K.L. and D.E. performed the protein pulldown studies and interpreted data. M.T. performed mESC knockdown and knockout experiments. J.S. and O. Kosmatchev did sample preparation and LC/MS analysis. B.H., S.S. and J.S. prepared MS standards. O. Kotljarova performed in vitro assays. B.S. synthesized oligonucleotides for protein capture. G.K. synthesized tandem mass tags, and S.M. provided mouse tissue samples. U.M. and H.L. constructed Tet expression plasmids. C.G.S. and M.V. performed Uhrf1/2 overexpression in HEK-293T cells. P.S. and D.S. provided plasmids and cell lines. M.M. and T.C. conceived and supervised the project, interpreted data and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Figures 1–12, Supplementary Tables 1–22 and Supplementary Notes 1–5. (PDF 2618 kb)
Supplementary Data Set 1
5hmU reader proteins identified from 4 independent pulldown experiments. (XLSX 126 kb)
Rights and permissions
About this article
Cite this article
Pfaffeneder, T., Spada, F., Wagner, M. et al. Tet oxidizes thymine to 5-hydroxymethyluracil in mouse embryonic stem cell DNA. Nat Chem Biol 10, 574–581 (2014). https://doi.org/10.1038/nchembio.1532
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.1532
This article is cited by
-
DNA modifications walk a fine line between epigenetics and mutagenesis
Nature Reviews Molecular Cell Biology (2023)
-
FANCD2 maintains replication fork stability during misincorporation of the DNA demethylation products 5-hydroxymethyl-2’-deoxycytidine and 5-hydroxymethyl-2’-deoxyuridine
Cell Death & Disease (2022)
-
Isoform-specific and ubiquitination dependent recruitment of Tet1 to replicating heterochromatin modulates methylcytosine oxidation
Nature Communications (2022)
-
RNA modifications: importance in immune cell biology and related diseases
Signal Transduction and Targeted Therapy (2022)
-
Dynamic changes in genomic 5-hydroxymethyluracil and N6-methyladenine levels in the Drosophila melanogaster life cycle and in response to different temperature conditions
Scientific Reports (2022)