Abstract
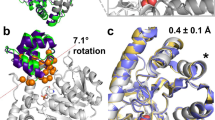
We emphasize the importance of dynamics and hydration for enzymatic catalysis and protein design by transplanting the active site from a haloalkane dehalogenase with high enantioselectivity to nonselective dehalogenase. Protein crystallography confirms that the active site geometry of the redesigned dehalogenase matches that of the target, but its enantioselectivity remains low. Time-dependent fluorescence shifts and computer simulations revealed that dynamics and hydration at the tunnel mouth differ substantially between the redesigned and target dehalogenase.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Damborsky, J. & Brezovsky, J. Curr. Opin. Chem. Biol. 19, 8–16 (2014).
Koehl, P. & Levitt, M. J. Mol. Biol. 293, 1161–1181 (1999).
Siegel, J.B. et al. Science 329, 309–313 (2010).
Röthlisberger, D. et al. Nature 453, 190–195 (2008).
Jiang, L. et al. Science 319, 1387–1391 (2008).
Kries, H., Blomberg, R. & Hilvert, D. Curr. Opin. Chem. Biol. 17, 221–228 (2013).
Khersonsky, O. et al. Proc. Natl. Acad. Sci. USA 109, 10358–10363 (2012).
Khare, S.D. et al. Nat. Chem. Biol. 8, 294–300 (2012).
Giger, L. et al. Nat. Chem. Biol. 9, 494–498 (2013).
Blomberg, R. et al. Nature 503, 418–421 (2013).
Sato, Y. et al. Appl. Environ. Microbiol. 71, 4372–4379 (2005).
Kulakova, A.N., Larkin, M.J. & Kulakov, L.A. Microbiology 143, 109–115 (1997).
Chovancová, E., Kosinski, J., Bujnicki, J.M. & Damborsky, J. Proteins 67, 305–316 (2007).
Prokop, Z. et al. Angew. Chem. Int. Edn Engl. 49, 6111–6115 (2010).
Eisenmesser, E.Z. et al. Nature 438, 117–121 (2005).
Henzler-Wildman, K. & Kern, D. Nature 450, 964–972 (2007).
Bhabha, G. et al. Science 332, 234–238 (2011).
Jimenez, R., Fleming, G.R., Kumar, P.V. & Maroncelli, M. Nature 369, 471–473 (1994).
Halle, B. & Nilsson, L. J. Phys. Chem. B 113, 8210–8213 (2009).
Jesenská, A. et al. J. Am. Chem. Soc. 131, 494–501 (2009).
Chang, C.-W. et al. Proc. Natl. Acad. Sci. USA 107, 2914–2919 (2010).
Jurkiewicz, P., Cwiklik, L., Jungwirth, P. & Hof, M. Biochimie 94, 26–32 (2012).
Nilsson, L. & Halle, B. Proc. Natl. Acad. Sci. USA 102, 13867–13872 (2005).
Baker, D. Protein Sci. 19, 1817–1819 (2010).
Chovancova, E. et al. PLoS Comput. Biol. 8, e1002708 (2012).
Sambrook, J. & Russell, D.W. Molecular Cloning: a Laboratory Manual. (Cold Spring Harbor Laboratory Press, New York, 2001).
Iwasaki, I., Utsumi, S. & Ozawa, T. Bull. Chem. Soc. Jpn. 25, 226 (1952).
Furse, K.E. & Corcelli, S.A. J. Phys. Chem. Lett. 1, 1813–1820 (2010).
Ducruix, A. & Giegé, R. Crystallization of Nucleic Acids and Proteins: a Practical Approach. (Oxford University Press, New York, 1999).
Teng, T.Y. J. Appl. Crystallogr. 23, 387–391 (1990).
Vagin, A. & Teplyakov, A. J. Appl. Crystallogr. 30, 1022–1025 (1997).
Murshudov, G.N., Vagin, A.A. & Dodson, E.J. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 (1997).
Sheldrick, G.M. Acta Crystallogr. A 64, 112–122 (2008).
Emsley, P., Lohkamp, B., Scott, W.G. & Cowtan, K. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Acknowledgements
We thank D. Baker (University of Washington) for helpful comments on the manuscript. This work was supported by the European Regional Development Fund (CZ.1.05/1.1.00/02.0123), the Czech Ministry of Education (LO1214), the Czech Science Foundation (P208/12/G016 to J.S. and M.H. and P503/12/0572 to J.D.) and the 'Employment of Best Young Scientists for International Cooperation Empowerment' program (CZ1.07/2.3.00/30.0037 to J.B.), financed by both the European Social Fund and the state budget of the Czech Republic. The Academy of Sciences is acknowledged for the Praemium Academie award (M.H.). CERIT Scientific Cloud is acknowledged for providing access to their computing facilities under the Center CERIT Scientific Cloud program (CZ.1.05/3.2.00/08.0144).
Author information
Authors and Affiliations
Contributions
J.S. and T.C. conducted fluorescence spectroscopy measurements. J.B. performed molecular modeling. T.K. and A.F. constructed the mutants. T.K. and Z.P. biochemically characterized the mutants. T.K., V.S. and R.C. conducted CD spectroscopy measurements. M.L. and I.K.S. determined the crystal structure. M.H. and J.D. conceived and supervised the project. J.S., J.B., T.K., M.H. and J.D. wrote the paper together.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Note, Supplementary Results, Supplementary Figures 1–11 and Supplementary Tables 1–9. (PDF 2175 kb)
Rights and permissions
About this article
Cite this article
Sykora, J., Brezovsky, J., Koudelakova, T. et al. Dynamics and hydration explain failed functional transformation in dehalogenase design. Nat Chem Biol 10, 428–430 (2014). https://doi.org/10.1038/nchembio.1502
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.1502
This article is cited by
-
Spatiotemporal multiscale molecular cavity visualization and visual analysis
Journal of Visualization (2020)
-
The role of protein dynamics in the evolution of new enzyme function
Nature Chemical Biology (2016)
-
Properties and biotechnological applications of natural and engineered haloalkane dehalogenases
Applied Microbiology and Biotechnology (2015)