Abstract
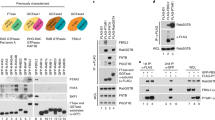
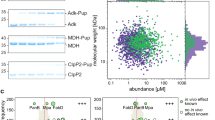
Protein prenylation is a widespread phenomenon in eukaryotic cells that affects many important signaling molecules. We describe the structure-guided design of engineered protein prenyltransferases and their universal synthetic substrate, biotin-geranylpyrophosphate. These new tools allowed us to detect femtomolar amounts of prenylatable proteins in cells and organs and to identify their cognate protein prenyltransferases. Using this approach, we analyzed the in vivo effects of protein prenyltransferase inhibitors. Whereas some of the inhibitors displayed the expected activities, others lacked in vivo activity or targeted a broader spectrum of prenyltransferases than previously believed. To quantitate the in vivo effect of the prenylation inhibitors, we profiled biotin-geranyl–tagged RabGTPases across the proteome by mass spectrometry. We also demonstrate that sites of active vesicular transport carry most of the RabGTPases. This approach enables a quantitative proteome-wide analysis of the regulation of protein prenylation and its modulation by therapeutic agents.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Walsh, C.T., Garneau-Tsodikova, S. & Gatto, G.J. Jr. Protein posttranslational modifications: the chemistry of proteome diversifications. Angew. Chem. Int. Edn Engl. 44, 7342–7372 (2005).
Meri, S. & Baumann, M. Proteomics: posttranslational modifications, immune responses and current analytical tools. Biomol. Eng. 18, 213–220 (2001).
Prescher, J.A. & Bertozzi, C.R. Chemistry in living systems. Nat. Chem. Biol. 1, 13–21 (2005).
Gelb, M.H. Protein prenylation, et cetera: signal transduction in two dimensions. Science 275, 1750–1751 (1997).
McTaggart, S.J. Isoprenylated proteins. Cell. Mol. Life Sci. 63, 255–267 (2006).
Casey, P.J. & Seabra, M.C. Protein prenyltransferases. J. Biol. Chem. 271, 5289–5292 (1996).
Maurer-Stroh, S. et al. Towards complete sets of farnesylated and geranylgeranylated proteins. PLoS Comput. Biol. 3, e66 (2007).
Armstrong, S.A., Brown, M.S., Goldstein, J.L. & Seabra, M.C. Preparation of recombinant Rab geranylgeranyltransferase and Rab escort proteins. Methods Enzymol. 257, 30–41 (1995).
Wherlock, M., Gampel, A., Futter, C. & Mellor, H. Farnesyltransferase inhibitors disrupt EGF receptor traffic through modulation of the RhoB GTPase. J. Cell Sci. 117, 3221–3231 (2004).
Greenwood, J., Steinman, L. & Zamvil, S.S. Statin therapy and autoimmune disease: from protein prenylation to immunomodulation. Nat. Rev. Immunol. 6, 358–370 (2006).
Konstantinopoulos, P.A., Karamouzis, M.V. & Papavassiliou, A.G. Post-translational modifications and regulation of the RAS superfamily of GTPases as anticancer targets. Nat. Rev. Drug Discov. 6, 541–555 (2007).
Denoyelle, C. et al. Molecular mechanism of the anti-cancer activity of cerivastatin, an inhibitor of HMG-CoA reductase, on aggressive human breast cancer cells. Cell. Signal. 15, 327–338 (2003).
Lackner, M.R. et al. Chemical genetics identifies Rab geranylgeranyl transferase as an apoptotic target of farnesyl transferase inhibitors. Cancer Cell 7, 325–336 (2005).
Watanabe, M. et al. Inhibitors of protein geranylgeranyltransferase I and Rab geranylgeranyltransferase identified from a library of allenoate-derived compounds. J. Biol. Chem. 283, 9571–9579 (2008).
Hancock, J.F. Reticulocyte lysate assay for in vitro translation and posttranslational modification of Ras proteins. Methods Enzymol. 255, 60–65 (1995).
Peter, M., Chavrier, P., Nigg, E.A. & Zerial, M. Isoprenylation of rab proteins on structurally distinct cysteine motifs. J. Cell Sci. 102, 857–865 (1992).
Benetka, W., Koranda, M., Maurer-Stroh, S., Pittner, F. & Eisenhaber, F. Farnesylation or geranylgeranylation? Efficient assays for testing protein prenylation in vitro and in vivo. BMC Biochem. 7, 6 (2006).
Nguyen, U.T. et al. Exploiting the substrate tolerance of farnesyltransferase for site-selective protein derivatization. ChemBioChem 8, 408–423 (2007).
Kho, Y. et al. A tagging-via-substrate technology for detection and proteomics of farnesylated proteins. Proc. Natl. Acad. Sci. USA 101, 12479–12484 (2004).
Dursina, B. et al. Identification and specificity profiling of protein prenyltransferase inhibitors using new fluorescent phosphoisoprenoids. J. Am. Chem. Soc. 128, 2822–2835 (2006).
Baron, R. et al. RhoB prenylation is driven by the three carboxyl-terminal amino acids of the protein: evidenced in vivo by an anti-farnesyl cysteine antibody. Proc. Natl. Acad. Sci. USA 97, 11626–11631 (2000).
Troutman, J.M., Roberts, M.J., Andres, D.A. & Spielmann, H.P. Tools to analyze protein farnesylation in cells. Bioconjug. Chem. 16, 1209–1217 (2005).
Lane, K.T. & Beese, L.S. Thematic review series: lipid posttranslational modifications. Structural biology of protein farnesyltransferase and geranylgeranyltransferase type I. J. Lipid Res. 47, 681–699 (2006).
Turek-Etienne, T.C., Strickland, C.L. & Distefano, M.D. Biochemical and structural studies with prenyl diphosphate analogues provide insights into isoprenoid recognition by protein farnesyl transferase. Biochemistry 42, 3716–3724 (2003).
Guo, Z. et al. Structures of RabGGTase-substrate/product complexes provide insights into the evolution of protein prenylation. EMBO J. 27, 2444–2456 (2008).
Reigard, S.A. et al. Interplay of isoprenoid and peptide substrate specificity in protein farnesyltransferase. Biochemistry 44, 11214–11223 (2005).
Krzysiak, A.J. et al. Combinatorial modulation of protein prenylation. ACS Chem. Biol. 2, 385–389 (2007).
Cassidy, P.B., Dolence, J.M. & Poulter, C.D. Continuous fluorescence assay for protein prenyltransferases. Methods Enzymol. 250, 30–43 (1995).
Cox, A.D. et al. The CAAX peptidomimetic compound B581 specifically blocks farnesylated, but not geranylgeranylated or myristylated, oncogenic ras signaling and transformation. J. Biol. Chem. 269, 19203–19206 (1994).
McGuire, T.F., Qian, Y., Vogt, A., Hamilton, A.D. & Sebti, S.M. Platelet-derived growth factor receptor tyrosine phosphorylation requires protein geranylgeranylation but not farnesylation. J. Biol. Chem. 271, 27402–27407 (1996).
Delahunty, C.M. & Yates, J.R. III . MudPIT: multidimensional protein identification technology. Biotechniques 43 563, 565, 567 (2007).
Lutcke, A. et al. Rab17, a novel small GTPase, is specific for epithelial cells and is induced during cell polarization. J. Cell Biol. 121, 553–564 (1993).
Florens, L. et al. A proteomic view of the Plasmodium falciparum life cycle. Nature 419, 520–526 (2002).
Zybailov, B., Coleman, M.K., Florens, L. & Washburn, M.P. Correlation of relative abundance ratios derived from peptide ion chromatograms and spectrum counting for quantitative proteomic analysis using stable isotope labeling. Anal. Chem. 77, 6218–6224 (2005).
Chen, B.C., Sundeen, J.E., Guo, P., Bednarz, M.S. & Zhao, R. Novel triethylsilane mediated reductive N-alkylation of amines: improved synthesis of 1-(4-imidazolyl)methyl-4-sulfonylbenzodiazepines new farnesyltransferase inhibitors. Tetrahedron Lett. 42, 1245–1246 (2001).
Kabsch, W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 26, 795–800 (1993).
McCoy, A.J., Grosse-Kunstleve, R.W., Storoni, L.C. & Read, R.J. Likelihood-enhanced fast translation functions. Acta Crystallogr. D Biol. Crystallogr. 61, 458–464 (2005).
Murshudov, G.N., Vagin, A.A. & Dodson, E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 (1997).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Schuttelkopf, A.W. & van Aalten, D.M. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. D Biol. Crystallogr. 60, 1355–1363 (2004).
Acknowledgements
This work was supported in part by grant Deutsche Forschungsgemeinschaft AL 484/7-2 to K.A. and grant Sonderforschungsbereich 642 to K.A., D.W., R.S.G. and H.W. U.T.T.N. was supported by the predoctoral fellowship of Fonds der chemischen Industrie. R.S.B. and C. Deraeve thank the Alexander von Humboldt Stiftung for a scholarship. We thank R. Heuann (Ruhr-Universität Bochum) for supplying mouse brains. The authors gratefully acknowledge M. Terbeck, A. Sander, T. Rogowsky, S. Thuns and N. Lupilova for excellent technical assistance. The authors are very grateful to T. Bergbrede and the Dortmund Protein Facility at the Max Planck Institute. The use of beamlines at the Swiss Light Source (Paul Scherrrer Institute) and the help of the X-ray communities at the Max Planck Institute of Molecular Physiology and the Max Planck Insitute of Medical Research with data collection is gratefully acknowledged. We thank A. Barnekow (University of Münster) for the generous gift of Rab6A antibody.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7, Supplementary Tables 1 and 2, and Supplementary Methods (PDF 2405 kb)
Rights and permissions
About this article
Cite this article
Nguyen, U., Guo, Z., Delon, C. et al. Analysis of the eukaryotic prenylome by isoprenoid affinity tagging. Nat Chem Biol 5, 227–235 (2009). https://doi.org/10.1038/nchembio.149
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.149
This article is cited by
-
TET2 deficiency sensitizes tumor cells to statins by reducing HMGCS1 expression
Oncogene (2022)
-
Metabolic labeling with an alkyne probe reveals similarities and differences in the prenylomes of several brain-derived cell lines and primary cells
Scientific Reports (2021)
-
Proteome-wide analysis of protein lipidation using chemical probes: in-gel fluorescence visualization, identification and quantification of N-myristoylation, N- and S-acylation, O-cholesterylation, S-farnesylation and S-geranylgeranylation
Nature Protocols (2021)
-
Statin-mediated disruption of Rho GTPase prenylation and activity inhibits respiratory syncytial virus infection
Communications Biology (2021)
-
Dual chemical probes enable quantitative system-wide analysis of protein prenylation and prenylation dynamics
Nature Chemistry (2019)