Abstract
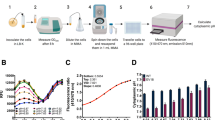
The widely conserved multiple antibiotic resistance regulator (MarR) family of transcription factors modulates bacterial detoxification in response to diverse antibiotics, toxic chemicals or both. The natural inducer for Escherichia coli MarR, the prototypical transcription repressor within this family, remains unknown. Here we show that copper signaling potentiates MarR derepression in E. coli. Copper(II) oxidizes a cysteine residue (Cys80) on MarR to generate disulfide bonds between two MarR dimers, thereby inducing tetramer formation and the dissociation of MarR from its cognate promoter DNA. We further discovered that salicylate, a putative MarR inducer, and the clinically important bactericidal antibiotics norfloxacin and ampicillin all stimulate intracellular copper elevation, most likely through oxidative impairment of copper-dependent envelope proteins, including NADH dehydrogenase-2. This membrane-associated copper oxidation and liberation process derepresses MarR, causing increased bacterial antibiotic resistance. Our study reveals that this bacterial transcription regulator senses copper(II) as a natural signal to cope with stress caused by antibiotics or the environment.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Martin, R.G., Nyantakyi, P.S. & Rosner, J.L. Regulation of the multiple antibiotic resistance (mar) regulon by marORA sequences in Escherichia coli. J. Bacteriol. 177, 4176–4178 (1995).
Alekshun, M.N. & Levy, S.B. Regulation of chromosomally mediated multiple antibiotic resistance: the mar regulon. Antimicrob. Agents Chemother. 41, 2067–2075 (1997).
Perera, I.C. & Grove, A. Molecular mechanisms of ligand-mediated attenuation of DNA binding by MarR family transcriptional regulators. J. Mol. Cell Biol. 2, 243–254 (2010).
Cohen, S.P., Levy, S.B., Foulds, J. & Rosner, J.L. Salicylate induction of antibiotic-resistance in Escherichia coli: activation of the mar operon and a mar-independent pathway. J. Bacteriol. 175, 7856–7862 (1993).
Alekshun, M.N. & Levy, S.B. The mar regulon: multiple resistance to antibiotics and other toxic chemicals. Trends Microbiol. 7, 410–413 (1999).
Martin, R.G. & Rosner, J.L. Binding of purified multiple antibiotic-resistance repressor protein (MarR) to mar operator sequences. Proc. Natl. Acad. Sci. USA 92, 5456–5460 (1995).
Alekshun, M.N. & Levy, S.B. Alteration of the repressor activity of MarR, the negative regulator of the Escherichia coli marRAB locus, by multiple chemicals in vitro. J. Bacteriol. 181, 4669–4672 (1999).
Price, C.T.D., Lee, I.R. & Gustafson, J.E. The effects of salicylate on bacteria. Int. J. Biochem. Cell Biol. 32, 1029–1043 (2000).
Nathan, C. & Cunningham-Bussel, A. Beyond oxidative stress: an immunologist's guide to reactive oxygen species. Nat. Rev. Immunol. 13, 349–361 (2013).
Kohanski, M.A., Dwyer, D.J., Hayete, B., Lawrence, C.A. & Collins, J.J. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130, 797–810 (2007).
Dwyer, D.J., Kohanski, M.A. & Collins, J.J. Role of reactive oxygen species in antibiotic action and resistance. Curr. Opin. Microbiol. 12, 482–489 (2009).
Liu, Y. & Imlay, J.A. Cell death from antibiotics without the involvement of reactive oxygen species. Science 339, 1210–1213 (2013).
Keren, I., Wu, Y., Inocencio, J., Mulcahy, L.R. & Lewis, K. Killing by bactericidal antibiotics does not depend on reactive oxygen species. Science 339, 1213–1216 (2013).
D'Autréaux, B. & Toledano, M.B. ROS as signalling molecules: mechanisms that generate specificity in ROS homeostasis. Nat. Rev. Mol. Cell Biol. 8, 813–824 (2007).
Pomposiello, P.J. & Demple, B. Redox-operated genetic switches: the SoxR and OxyR transcription factors. Trends Biotechnol. 19, 109–114 (2001).
Chen, P.R. et al. An oxidation-sensing mechanism is used by the global regulator MgrA in Staphylococcus aureus. Nat. Chem. Biol. 2, 591–595 (2006).
Lee, J.-W. & Helmann, J.D. The PerR transcription factor senses H2O2 by metal-catalysed histidine oxidation. Nature 440, 363–367 (2006).
Kohanski, M.A., DePristo, M.A. & Collins, J.J. Sublethal antibiotic treatment leads to multidrug resistance via radical-induced mutagenesis. Mol. Cell 37, 311–320 (2010).
Martin, R.G., Jair, K.W., Wolf, R.E. & Rosner, J.L. Autoactivation of the marRAB multiple antibiotic resistance operon by the MarA transcriptional activator in Escherichia coli. J. Bacteriol. 178, 2216–2223 (1996).
Uversky, V.N., Winter, S. & Löber, G. Self-association of 8-anilino-1-naphthalene-sulfonate molecules: spectroscopic characterization and application to the investigation of protein folding. Biochim. Biophys. Acta 1388, 133–142 (1998).
Xiao, Z., Loughlin, F., George, G.N., Howlett, G.J. & Wedd, A.G. C-terminal domain of the membrane copper transporter Ctr1 from Saccharomyces cerevisiae binds four Cu(I) ions as a cuprous-thiolate polynuclear cluster: sub-femtomolar Cu(I) affinity of three proteins involved in copper trafficking. J. Am. Chem. Soc. 126, 3081–3090 (2004).
Riddles, P.W., Blakeley, R.L. & Zerner, B. Reassessment of Ellman's reagent. Methods Enzymol. 91, 49–60 (1983).
Ding, X., Xie, H. & Kang, Y.J. The significance of copper chelators in clinical and experimental application. J. Nutr. Biochem. 22, 301–310 (2011).
Alekshun, M.N., Levy, S.B., Mealy, T.R., Seaton, B.A. & Head, J.F. The crystal structure of MarR, a regulator of multiple antibiotic resistance, at 2.3 Å resolution. Nat. Struct. Biol. 8, 710–714 (2001).
Hong, M., Fuangthong, M., Helmann, J.D. & Brennan, R.G. Structure of an OhrR–ohrA operator complex reveals the DNA binding mechanism of the MarR family. Mol. Cell 20, 131–141 (2005).
Kumarevel, T., Tanaka, T., Umehara, T. & Yokoyama, S.S.T. 1710–DNA complex crystal structure reveals the DNA binding mechanism of the MarR family of regulators. Nucleic Acids Res. 37, 4723–4735 (2009).
Dolan, K.T., Duguid, E.M. & He, C. Crystal structures of SlyA protein, a master virulence regulator of Salmonella, in free and DNA-bound states. J. Biol. Chem. 286, 22178–22185 (2011).
Dwyer, D.J., Kohanski, M.A., Hayete, B. & Collins, J.J. Gyrase inhibitors induce an oxidative damage cellular death pathway in Escherichia coli. Mol. Syst. Biol. 3, 91 (2007).
Zeng, L., Miller, E.W., Pralle, A., Isacoff, E.Y. & Chang, C.J. A selective turn-on fluorescent sensor for imaging copper in living cells. J. Am. Chem. Soc. 128, 10–11 (2006).
Kohanski, M.A., Dwyer, D.J., Wierzbowski, J., Cottarel, G. & Collins, J.J. Mistranslation of membrane proteins and two-component system activation trigger antibiotic-mediated cell death. Cell 135, 679–690 (2008).
Kershaw, C.J., Brown, N.L., Constantinidou, C., Patel, M.D. & Hobman, J.L. The expression profile of Escherichia coli K-12 in response to minimal, optimal and excess copper concentrations. Microbiology 151, 1187–1198 (2005).
Rodriguez-Montelongo, L., de la Cruz-Rodriguez, L.C., Farías, R.N. & Massa, E.M. Membrane-associated redox cycling of copper mediates hydroperoxide toxicity in. Escherichia coli. Biochim. Biophys. Acta 1144, 77–84 (1993).
Rodríguez-Montelongo, L.,, R.N. & Massa, E.M. Sites of electron transfer to membrane-bound copper and hydroperoxide-induced damage in the respiratory chain of Escherichia coli. Arch. Biochem. Biophys. 323, 19–26 (1995).
Walling, C. Fenton's reagent revisited. Acc. Chem. Res. 8, 125–131 (1975).
Patikarnmonthon, N. et al. Copper ions potentiate organic hydroperoxide and hydrogen peroxide toxicity through different mechanisms in Xanthomonas campestris pv. campestris. FEMS Microbiol. Lett. 313, 75–80 (2010).
Rapisarda, V.A. et al. Evidence for Cu(I)-thiolate ligation and prediction of a putative copper-binding site in the Escherichia coli NADH dehydrogenase-2. Arch. Biochem. Biophys. 405, 87–94 (2002).
Rodríguez-Montelongo, L., Volentini, S.I., Farías, R.N., Massa, E.M. & Rapisarda, V.A. The Cu(II)-reductase NADH dehydrogenase-2 of Escherichia coli improves the bacterial growth in extreme copper concentrations and increases the resistance to the damage caused by copper and hydroperoxide. Arch. Biochem. Biophys. 451, 1–7 (2006).
Zhao, B.S. et al. A highly selective fluorescent probe for visualization of organic hydroperoxides in living cells. J. Am. Chem. Soc. 132, 17065–17067 (2010).
Domain, F., Bina, X.R. & Levy, S.B. Transketolase A, an enzyme in central metabolism, derepresses the marRAB multiple antibiotic resistance operon of Escherichia coli by interaction with MarR. Mol. Microbiol. 66, 383–394 (2007).
Frawley, E.R. et al. Iron and citrate export by a major facilitator superfamily pump regulates metabolism and stress resistance in Salmonella Typhimurium. Proc. Natl. Acad. Sci. USA 110, 12054–12059 (2013).
Sun, F. et al. Quorum-sensing agr mediates bacterial oxidation response via an intramolecular disulfide redox switch in the response regulator AgrA. Proc. Natl. Acad. Sci. USA 109, 9095–9100 (2012).
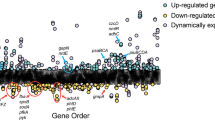
Deng, X. et al. Proteome-wide quantification and characterization of oxidation-sensitive cysteines in pathogenic bacteria. Cell Host Microbe 13, 358–370 (2013).
Winter, J., Ilbert, M., Graf, P.C.F., Özcelik, D. & Jakob, U. Bleach activates a redox-regulated chaperone by oxidative protein unfolding. Cell 135, 691–701 (2008).
Hiniker, A., Collet, J.-F. & Bardwell, J.C.A. Copper stress causes an in vivo requirement for the Escherichia coli disulfide isomerase DsbC. J. Biol. Chem. 280, 33785–33791 (2005).
Meloni, G. et al. Metal swap between Zn7-metallothionein-3 and amyloid-β-Cu protects against amyloid-β toxicity. Nat. Chem. Biol. 4, 366–372 (2008).
Battye, T.G.G., Kontogiannis, L., Johnson, O., Powell, H.R. & Leslie, A.G.W. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D Biol. Crystallogr. 67, 271–281 (2011).
Winn, M.D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 (2011).
McCoy, A.J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Murshudov, G.N., Vagin, A.A. & Dodson, E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 (1997).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Laskowski, R.A., MacArthur, M.W., Moss, D.S. & Thornton, J.M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 (1993).
Kleywegt, G.J. Use of non-crystallographic symmetry in protein structure refinement. Acta Crystallogr. D Biol. Crystallogr. 52, 842–857 (1996).
Painter, J. & Merritt, E.A. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr. D Biol. Crystallogr. 62, 439–450 (2006).
Chen, V.B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010).
Livak, K.J. & Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408 (2001).
Acknowledgements
We thank J. Rosner (National Institute of Diabetes and Digestive and Kidney Diseases, US National Institutes of Health) for providing the strains of M2073 (WT E. coli) and M2076 (marRAB mutant strains) bearing the marR::lacZ reporter, J.J. Collins (Department of Biomedical Engineering, Boston University) for providing the pL(lexO)-GFP reporter, C.J. Chang (College of Chemistry, University of California–Berkeley) for providing the Cu(I)-specific fluorescent probe CS-1 and S.F. Reichard for editing. The E. coli WT (K12) strain (BW25113) and all of the single-gene deletion mutants were obtained from the National BioResource Project (National Institute of Genetics, Japan). We also thank the staff members of the Shanghai Synchrotron Radiation Facility and the Beijing Synchrotron Radiation Facility. This work was supported by research grants from the National Basic Research Foundation of China (2010CB912302 and 2012CB917301 to P.R.C.; 2011CB809103 to C.H.), the National Natural Science Foundation of China (21225206, 91013005 and 21001010 to P.R.C.), the US National Science Foundation (CHE-1213598 to C.H.) and the E-Institutes of Shanghai Municipal Education Commission (project number E09013 to C.H. and P.R.C.).
Author information
Authors and Affiliations
Contributions
Z.H. performed the biochemical studies and participated in the structure studies. H.L. determined the crystal structures of copper(II)-oxidized MarR5CS(80C) and reduced MarRC80S. R.Z., J.Z. and X.C. helped with the biochemical and/or structure experiments. D.Z., B.S.Z. and S.Z. performed the experiments for OHP detection using OHSer. J.C. synthesized the Cu(I)-specific fluorescent probe CS-1. P.R.C. and C.H. conceived the study, designed experiments, interpreted data and wrote the manuscript with input from all of the authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Tables 1–6 and Supplementary Figures 1–24. (PDF 4517 kb)
Rights and permissions
About this article
Cite this article
Hao, Z., Lou, H., Zhu, R. et al. The multiple antibiotic resistance regulator MarR is a copper sensor in Escherichia coli. Nat Chem Biol 10, 21–28 (2014). https://doi.org/10.1038/nchembio.1380
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.1380
This article is cited by
-
A novel Escherichia coli cell–based bioreporter for quantification of salicylic acid in cosmetics
Applied Microbiology and Biotechnology (2024)
-
Copper and nanostructured anatase rutile and carbon coatings induce adaptive antibiotic resistance
AMB Express (2022)
-
Combining CRISPRi and metabolomics for functional annotation of compound libraries
Nature Chemical Biology (2022)
-
EpsRAc is a copper-sensing MarR family transcriptional repressor from Acidithiobacillus caldus
Applied Microbiology and Biotechnology (2022)
-
The MarR family regulator OsbR controls oxidative stress response, anaerobic nitrate respiration, and biofilm formation in Chromobacterium violaceum
BMC Microbiology (2021)