Abstract
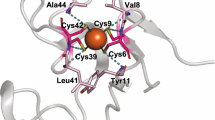
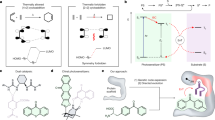
Emulating functions of natural enzymes in man-made constructs has proven challenging. Here we describe a man-made protein platform that reproduces many of the diverse functions of natural oxidoreductases without importing the complex and obscure interactions common to natural proteins. Our design is founded on an elementary, structurally stable 4-α-helix protein monomer with a minimalist interior malleable enough to accommodate various light- and redox-active cofactors and with an exterior tolerating extensive charge patterning for modulation of redox cofactor potentials and environmental interactions. Despite its modest size, the construct offers several independent domains for functional engineering that targets diverse natural activities, including dioxygen binding and superoxide and peroxide generation, interprotein electron transfer to natural cytochrome c and light-activated intraprotein energy transfer and charge separation approximating the core reactions of photosynthesis, cryptochrome and photolyase. The highly stable, readily expressible and biocompatible characteristics of these open-ended designs promise development of practical in vitro and in vivo applications.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Change history
11 December 2013
In the version of this article initially published, the US Department of Energy Office of Basic Energy Sciences, Energy Frontier Research Center grant number was incorrect. The correct number is DE-SC 0001035. The error has been corrected in the HTML and PDF versions of the article.
References
Fischer, E. Nobel Lectures, Chemistry 1901–1921 Vol.1 (ed. Nobelstiftelsen) 21–35 (Elsevier, Amsterdam, 1966).
Brustad, E.M. & Arnold, F.H. Optimizing non-natural protein function with directed evolution. Curr. Opin. Chem. Biol. 15, 201–210 (2011).
Baker, D. An exciting but challenging road ahead for computational enzyme design. Protein Sci. 19, 1817–1819 (2010).
Prabhulkar, S., Tian, H., Wang, X.T., Zhu, J.J. & Li, C.Z. Engineered proteins: redox properties and their applications. Antioxid. Redox Signal. 17, 1796–1822 (2012).
Edelman, G.M. & Gally, J.A. Degeneracy and complexity in biological systems. Proc. Natl. Acad. Sci. USA 98, 13763–13768 (2001).
Lichtenstein, B.R. et al. Engineering oxidoreductases: maquette proteins designed from scratch. Biochem. Soc. Trans. 40, 561–566 (2012).
Regan, L. & DeGrado, W.F. Characterization of a helical protein designed from first principles. Science 241, 976–978 (1988).
Page, C.C., Moser, C.C., Chen, X. & Dutton, P.L. Natural engineering principles of electron tunnelling in biological oxidation-reduction. Nature 402, 47–52 (1999).
Robertson, D.E. et al. Design and synthesis of multi-heme proteins. Nature 368, 425–432 (1994).
Sharp, R.E., Moser, C.C., Rabanal, F. & Dutton, P.L. Design, synthesis, and characterization of a photoactivatable flavocytochrome molecular maquette. Proc. Natl. Acad. Sci. USA 95, 10465–10470 (1998).
Shifman, J.M., Moser, C.C., Kalsbeck, W.A., Bocian, D.F. & Dutton, P.L. Functionalized de novo designed proteins: mechanism of proton coupling to oxidation/reduction in heme protein maquettes. Biochemistry 37, 16815–16827 (1998).
Grosset, A.M., Gibney, B.R., Rabanal, F., Moser, C.C. & Dutton, P.L. Proof of principle in a de novo designed protein maquette: an allosterically regulated, charge-activated conformational switch in a tetra-α-helix bundle. Biochemistry 40, 5474–5487 (2001).
Anderson, J.L.R., Koder, R.L., Moser, C.C. & Dutton, P.L. Controlling complexity and water penetration in functional de novo protein design. Biochem. Soc. Trans. 36, 1106–1111 (2008).
Koder, R.L. et al. Design and engineering of an O2 transport protein. Nature 458, 305–309 (2009).
Bender, G.M. et al. De novo design of a single-chain diphenylporphyrin metalloprotein. J. Am. Chem. Soc. 129, 10732–10740 (2007).
Ku, J. & Schultz, P.G. Alternate protein frameworks for molecular recognition. Proc. Natl. Acad. Sci. USA 92, 6552–6556 (1995).
Predki, P.F. & Regan, L. Redesigning the topology of a four-helix-bundle protein: monomeric Rop. Biochemistry 34, 9834–9839 (1995).
Westerlund, K. et al. Making a single-chain four-helix bundle for redox chemistry studies. Protein Eng. Des. Sel. 21, 645–652 (2008).
Huang, S.S., Gibney, B.R., Stayrook, S.E., Dutton, P.L. & Lewis, M. X-ray structure of a Maquette scaffold. J. Mol. Biol. 326, 1219–1225 (2003).
Aurora, R. & Rose, G.D. Helix capping. Protein Sci. 7, 21–38 (1998).
Koder, R.L. et al. Native like structure in designed four α-helix bundles driven by buried polar interactions. J. Am. Chem. Soc. 128, 14450–14451 (2006).
Caffrey, M.S. & Cusanovich, M.A. Site-specific mutagenesis studies of cytochromes c. Biochim. Biophys. Acta 1187, 277–288 (1994).
Davies, A.M. et al. Redesign of the interior hydrophilic region of mitochondrial cytochrome c by site-directed mutagenesis. Biochemistry 32, 5431–5435 (1993).
Trent, J.T., Hvitved, A.N. & Hargrove, M.S. A model for ligand binding to hexacoordinate hemoglobins. Biochemistry 40, 6155–6163 (2001).
Kiger, L. et al. Electron transfer function versus oxygen delivery: a comparative study for several hexacoordinated globins across the animal kingdom. PLoS ONE 6, e20478 (2011).
Hamdane, D. et al. Hyperthermal stability of neuroglobin and cytoglobin. FEBS J. 272, 2076–2084 (2005).
Fago, A., Mathews, A.J., Moens, L., Dewilde, S. & Brittain, T. The reaction of neuroglobin with potential redox protein partners cytochrome b5 and cytochrome c. FEBS Lett. 580, 4884–4888 (2006).
Margoliash, E. & Bosshard, H.R. Guided by electrostatics, a textbook protein comes of age. Trends Biochem. Sci. 8, 316–320 (1983).
Moser, C.C. & Dutton, P.L. Cytochrome c and c2 binding dynamics and electron transfer with photosynthetic reaction center protein and other integral membrane redox proteins. Biochemistry 27, 2450–2461 (1988).
Ku, H.H., Brunk, U.T. & Sohal, R.S. Relationship between mitochondrial superoxide and hydrogen-peroxide production and longevity of mammalian-species. Free Radic. Biol. Med. 15, 621–627 (1993).
Muir Wood, P. The redox potential of the system oxygen—superoxide. FEBS Lett. 44, 22–24 (1974).
Zhang, L., Andersen, E.M.E., Khajo, A., Magliozzo, R.S. & Koder, R.L. Dynamic factors affecting gaseous ligand binding in an artificial oxygen transport protein. Biochemistry 52, 447–455 (2013).
Cross, A.R., Parkinson, J.F. & Jones, O.T.G. Mechanism of the superoxide-producing oxidase of neutrophils—O2 is necessary for the fast reduction of cytochrome b245 by NADPH. Biochem. J. 226, 881–884 (1985).
Lin, C. et al. Association of flavin adenine-dinucleotide with the Arabidopsis blue-light receptor Cry1. Science 269, 968–970 (1995).
Aubert, C., Vos, M.H., Mathis, P., Eker, A.P. & Brettel, K. Intraprotein radical transfer during photoactivation of DNA photolyase. Nature 405, 586–590 (2000).
Muthiah, C., Ptaszek, M., Nguyen, T.M., Flack, K.M. & Lindsey, J.S. Two complementary routes to 7-substituted chlorins. Partial mimics of chlorophyll b. J. Org. Chem. 72, 7736–7749 (2007).
Moser, C.C., Anderson, J.L. & Dutton, P.L. Guidelines for tunneling in enzymes. Biochim. Biophys. Acta 1797, 1573–1586 (2010).
Moser, C.C., Keske, J.M., Warncke, K., Farid, R.S. & Dutton, P.L. Nature of biological electron transfer. Nature 355, 796–802 (1992).
Massey, V., Stankovich, M. & Hemmerich, P. Light-mediated reduction of flavoproteins with flavins as catalysts. Biochemistry 17, 1–8 (1978).
Grzyb, J. et al. Empirical and computational design of iron-sulfur cluster proteins. Biochim. Biophys. Acta 1817, 1256–1262 (2012).
Reedy, C.J. & Gibney, B.R. Heme protein assemblies. Chem. Rev. 104, 617–649 (2004).
Monien, B.H., Drepper, F., Sommerhalter, M., Lubitz, W. & Haehnel, W. Detection of heme oxygenase activity in a library of four-helix bundle proteins: towards the de novo synthesis of functional heme proteins. J. Mol. Biol. 371, 739–753 (2007).
Patel, S.C. & Hecht, M.H. Directed evolution of the peroxidase activity of a de novo–designed protein. Protein Eng. Des. Sel. 25, 445–452 (2012).
Smith, B.A. & Hecht, M.H. Novel proteins: from fold to function. Curr. Opin. Chem. Biol. 15, 421–426 (2011).
Fry, H.C., Lehmann, A., Saven, J.G., DeGrado, W.F. & Therien, M.J. Computational design and elaboration of a de novo heterotetrameric α-helical protein that selectively binds an emissive abiological (porphinato)zinc chromophore. J. Am. Chem. Soc. 132, 3997–4005 (2010).
Reig, A.J. et al. Alteration of the oxygen-dependent reactivity of de novo Due Ferri proteins. Nat. Chem. 4, 900–906 (2012).
Röthlisberger, D. et al. Kemp elimination catalysts by computational enzyme design. Nature 453, 190–195 (2008).
Miner, K.D. et al. A designed functional metalloenzyme that reduces O2 to H2O with over one thousand turnovers. Angew. Chem. Int. Ed. Engl. 51, 5589–5592 (2012).
Gibney, B.R., Mulholland, S.E., Rabanal, F. & Dutton, P.L. Ferredoxin and ferredoxin-heme maquettes. Proc. Natl. Acad. Sci. USA 93, 15041–15046 (1996).
Fisher, M.A., McKinley, K.L., Bradley, L.H., Viola, S.R. & Hecht, M.H. De novo designed proteins from a library of artificial sequences function in Escherichia coli and enable cell growth. PLoS ONE 6, e15364 (2011).
Berry, E.A. & Trumpower, B.L. Simultaneous determination of hemes a, b and c from pyridine hemochrome spectra. Anal. Biochem. 161, 1–15 (1987).
Acknowledgements
In this research, the US National Institutes of Health (NIH)–General Medical Institutes (RO1 GM 41048) funded the design and development of the maquette proteins A, B, F, G and H, including gene design, cloning, protein expression, purification and characterization; it also funded the thermal stability measurements using CD and demonstrations of control of oxygen binding and redox chemistry. Basic to these developments was NMR spectroscopy performed by M.A.S., K.G.V. and A.J.W., supported by NIH United States Public Health Service grants DK39806 and GM102477 to A.J.W. The US Department of Energy, Office of Basic Energy Sciences, Division of Materials Sciences and Engineering (DE-FG02-05ER46223) funded the synthesis and characterization of flavins and also the design, expression, purification and characterization of C, K, J protein maquettes promoting light-activated charge separation and oxidation-reduction using flavin and Zn- and Fe- tetrapyrroles as cofactors. The US Department of Energy Office of Basic Energy Sciences, Energy Frontier Research Center (PARC) (DE-SC 0001035 to P.L.D. and C.C.M.) funded development of light excitation energy transfer in maquettes (L and its mutants), synthesis and purification of Zn pyropheophoribide a and covalent attachments of Alexa Fluor to the maquettes. In this work, the synthetic chlorin ZnC was a generous gift from O. Mass and J.S. Lindsey at North Carolina State University.
Author information
Authors and Affiliations
Contributions
T.A.F. contributed to the design and characterization of maquettes A, E, J, H and B; G.K. designed, expressed, purified and characterized maquettes C, L, D, K and F and contributed to all of the experiments performed with these maquettes as well as the writing of the manuscript; L.A.S. performed the redox titrations, developed and characterized maquette G and performed CO and O2 kinetics on maquette A as well as assisted in the writing of the manuscript; B.R.L. contributed to the monomeric maquette design as well as experimental design and interpretation; M.M.S. designed and purified maquette I and measured superoxide production, low-temperature spectra and oxyferrous state kinetics; B.A.F. measured electron transfer from A to cytochrome c; C.B. synthesized and characterized flavomaquettes; N.M.E. performed synthetic chlorin binding affinity measurements and contributed to protein design; J.A.S. contributed to heme and Zn porphyrin binding affinities of C and J; Z.Z. contributed to protein design; B.M.D. contributed to experimental designs and manuscript writing; M.A.S., K.G.V. and A.J.W. contributed to the NMR characterization of maquettes; J.L.R.A. contributed to CO and O2 ligand kinetics for maquette A. C.C.M. designed and operated transient spectroscopy equipment for photolysis and light-induced electron transfer and contributed substantially to manuscript writing; P.L.D. conceived and designed experiments and contributed substantially to manuscript writing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Table 1 and Supplementary Figures 1–43. (PDF 4867 kb)
Rights and permissions
About this article
Cite this article
Farid, T., Kodali, G., Solomon, L. et al. Elementary tetrahelical protein design for diverse oxidoreductase functions. Nat Chem Biol 9, 826–833 (2013). https://doi.org/10.1038/nchembio.1362
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.1362
This article is cited by
-
Mn-porphyrins in a four-helix bundle participate in photo-induced electron transfer with a bacterial reaction center
Photosynthesis Research (2023)
-
De novo protein design of photochemical reaction centers
Nature Communications (2022)
-
A bound iron porphyrin is redox active in hybrid bacterial reaction centers modified to possess a four-helix bundle domain
Photochemical & Photobiological Sciences (2022)
-
Design of buried charged networks in artificial proteins
Nature Communications (2021)
-
Small-residue packing motifs modulate the structure and function of a minimal de novo membrane protein
Scientific Reports (2020)