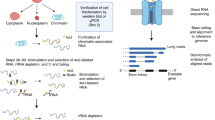
Abstract
Polyadenylation of mRNA leads to increased protein expression in response to diverse stimuli, but it is difficult to identify mRNAs that become polyadenylated in living cells. Here we describe a click chemistry–compatible nucleoside analog that is selectively incorporated into poly(A) tails of transcripts in cells. Next-generation sequencing of labeled mRNAs enables a transcriptome-wide profile of polyadenylation and provides insights into the mRNA sequence elements that are correlated with polyadenylation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Weill, L., Belloc, E., Bava, F.A. & Mendez, R. Nat. Struct. Mol. Biol. 19, 577–585 (2012).
Meijer, H.A. et al. Nucleic Acids Res. 35, e132 (2007).
Novoa, I., Gallego, J., Ferreira, P.G. & Mendez, R. Nat. Cell Biol. 12, 447–456 (2010).
Graindorge, A., Thuret, R., Pollet, N., Osborne, H.B. & Audic, Y. Nucleic Acids Res. 34, 986–995 (2006).
Beilharz, T.H. & Preiss, T. Methods 48, 294–300 (2009).
Meijer, H.A. & de Moor, C.H. Methods Mol. Biol. 703, 123–135 (2011).
Martin, G., Keller, W. & Doublie, S. EMBO J. 19, 4193–4203 (2000).
Chen, L.S. & Sheppard, T.L. J. Biol. Chem. 279, 40405–40411 (2004).
Martin, G., Moglich, A., Keller, W. & Doublie, S. J. Mol. Biol. 341, 911–925 (2004).
Horowitz, B., Goldfinger, B.A. & Marmur, J. Arch. Biochem. Biophys. 172, 143–148 (1976).
Jao, C.Y. & Salic, A. Proc. Natl. Acad. Sci. USA 105, 15779–15784 (2008).
Radford, H.E., Meijer, H.A. & de Moor, C.H. Biochim. Biophys. Acta 1779, 217–229 (2008).
Schorderet-Slatkine, S. Cell Differ. 1, 179–189 (1972).
de Moor, C.H., Meijer, H. & Lissenden, S. Semin. Cell Dev. Biol. 16, 49–58 (2005).
de Moor, C.H. & Richter, J.D. Mol. Cell Biol. 17, 6419–6426 (1997).
Sheets, M.D., Fox, C.A., Hunt, T., Vande Woude, G. & Wickens, M. Genes Dev. 8, 926–938 (1994).
Charlesworth, A., Cox, L.L. & MacNicol, A.M. J. Biol. Chem. 279, 17650–17659 (2004).
Paris, J. & Philippe, M. Dev. Biol. 140, 221–224 (1990).
Culp, P.A. & Musci, T.J. Dev. Biol. 193, 63–76 (1998).
Rassa, J.C., Wilson, G.M., Brewer, G.A. & Parks, G.D. Virology 274, 438–449 (2000).
MacNicol, M.C. & MacNicol, A.M. Mol. Reprod. Dev. 77, 662–669 (2010).
Cohen, M.S., Bas Orth, C., Kim, H.J., Jeon, N.L. & Jaffrey, S.R. Proc. Natl. Acad. Sci. USA 108, 11246–11251 (2011).
Krek, W. & DeCaprio, J.A. Methods Enzymol. 254, 114–124 (1995).
Groisman, I. et al. Cell 103, 435–447 (2000).
Cohen, S., Au, S. & Pante, N. J. Vis. Exp. http://dx.doi.org/10.3791/1106 (23 February 2009).
Jiang, H. & Wong, W.H. Bioinformatics 24, 2395–2396 (2008).
Anders, S. & Huber, W. Genome Biol. 11, R106 (2010).
Huang da, W., Sherman, B.T. & Lempicki, R.A. Nat. Protoc. 4, 44–57 (2009).
Huang da, W., Sherman, B.T. & Lempicki, R.A. Nucleic Acids Res. 37, 1–13 (2009).
Markham, N.R. & Zuker, M. Methods Mol. Biol. 453, 3–31 (2008).
Acknowledgements
We thank members of the Jaffrey lab for helpful comments and suggestions. We thank J. Richter (University of Massachusetts Medical School) for the antibody to CPEB. We gratefully acknowledge C. Bracken and N. Svensen for assistance with NMR spectroscopy. This work was supported by US National Institutes of Health–National Institute on Drug Abuse grant T32DA007274 (D.C. and M.C.) and US National Institute of Neurological Disorders and Stroke grant R01NS56306 (S.R.J.).
Author information
Authors and Affiliations
Contributions
D.C. and S.R.J. designed experiments, analyzed the data and wrote the manuscript; D.C. performed the experiments; C.E.S. performed oocyte injection experiments; I.S. and C.S.L. performed bioinformatics analyses; M.C. designed and synthesized 2-ethynyl adenosine and EAMP and wrote portions of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Figures 1–16, Supplementary Note and Supplementary Tables 1–18. (PDF 34944 kb)
Supplementary Data Set 1
List of EA-trapped and EA-depleted transcripts (XLSX 87 kb)
Rights and permissions
About this article
Cite this article
Curanovic, D., Cohen, M., Singh, I. et al. Global profiling of stimulus-induced polyadenylation in cells using a poly(A) trap. Nat Chem Biol 9, 671–673 (2013). https://doi.org/10.1038/nchembio.1334
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.1334
This article is cited by
-
Total RNA Synthesis and its Covalent Labeling Innovation
Topics in Current Chemistry (2022)
-
Comparisons among rainbow trout, Oncorhynchus mykiss, populations of maternal transcript profile associated with egg viability
BMC Genomics (2021)
-
Transcriptome analysis of egg viability in rainbow trout, Oncorhynchus mykiss
BMC Genomics (2019)
-
An Integrated Chemical Proteomics Approach for Quantitative Profiling of Intracellular ADP-Ribosylation
Scientific Reports (2019)
-
Musashi 1 regulates the timing and extent of meiotic mRNA translational activation by promoting the use of specific CPEs
Nature Structural & Molecular Biology (2017)