Abstract
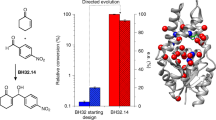
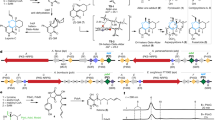
Evolutionary advances are often fueled by unanticipated innovation. Directed evolution of a computationally designed enzyme suggests that pronounced molecular changes can also drive the optimization of primitive protein active sites. The specific activity of an artificial retro-aldolase was boosted >4,400-fold by random mutagenesis and screening, affording catalytic efficiencies approaching those of natural enzymes. However, structural and mechanistic studies reveal that the engineered catalytic apparatus, consisting of a reactive lysine and an ordered water molecule, was unexpectedly abandoned in favor of a new lysine residue in a substrate-binding pocket created during the optimization process. Structures of the initial in silico design, a mechanistically promiscuous intermediate and one of the most evolved variants highlight the importance of loop mobility and supporting functional groups in the emergence of the new catalytic center. Such internal competition between alternative reactive sites may have characterized the early evolution of many natural enzymes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Bolon, D.N. & Mayo, S.L. Enzyme-like proteins by computational design. Proc. Natl. Acad. Sci. USA 98, 14274–14279 (2001).
Jiang, L. et al. De novo computational design of retro-aldol enzymes. Science 319, 1387–1391 (2008).
Röthlisberger, D. et al. Kemp elimination catalysts by computational enzyme design. Nature 453, 190–195 (2008).
Privett, H.K. et al. Iterative approach to computational enzyme design. Proc. Natl. Acad. Sci. USA 109, 3790–3795 (2012).
Siegel, J.B. et al. Computational design of an enzyme catalyst for a stereoselective bimolecular Diels-Alder reaction. Science 329, 309–313 (2010).
Kries, H., Blomberg, R. & Hilvert, D. De novo enzymes by computational design. Curr. Opin. Chem. Biol. 17, 221–228 (2013).
Althoff, E.A. et al. Robust design and optimization of retroaldol enzymes. Protein Sci. 21, 717–726 (2012).
Khersonsky, O. et al. Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59. Proc. Natl. Acad. Sci. USA 109, 10358–10363 (2012).
Gefflaut, T., Blonski, C., Perie, J. & Willson, M.l. Class I aldolases: substrate specificity, mechanism, inhibitors and structural aspects. Prog. Biophys. Mol. Biol. 63, 301–340 (1995).
Lassila, J.K., Baker, D. & Herschlag, D. Origins of catalysis by computationally designed retroaldolase enzymes. Proc. Natl. Acad. Sci. USA 107, 4937–4942 (2010).
Wang, L. et al. Structural analyses of covalent enzyme-substrate analog complexes reveal strengths and limitations of de novo enzyme design. J. Mol. Biol. 415, 615–625 (2012).
Richter, F., Leaver-Fay, A., Khare, S.D., Bjelic, S. & Baker, D. De novo enzyme design using Rosetta3. PLoS ONE 6, e19230 (2011).
Tagaki, W. & Yamamoto, H. Polyamino-β-cyclodextrin as a model of aldolase. Tetrahedr. Lett. 32, 1207–1208 (1991).
Tanaka, F., Fuller, R. & Barbas, C.F. III. Development of small designer aldolase enzymes: catalytic activity, folding, and substrate specificity. Biochemistry 44, 7583–7592 (2005).
Müller, M.M., Windsor, M.A., Pomerantz, W.C., Gellman, S.H. & Hilvert, D. A rationally designed aldolase foldamer. Angew. Chem. Int. Ed. Engl. 48, 922–925 (2009).
Wörsdörfer, B., Henning, L., Obexer, R. & Hilvert, D. Harnessing protein symmetry for enzyme design. ACS Catal. 2, 982–985 (2012).
Wymer, N. et al. Directed evolution of a new catalytic site in 2-keto-3-deoxy-6-phosphogluconate aldolase from Escherichia coli. Structure 9, 1–9 (2001).
Heine, A. et al. Observation of covalent intermediates in an enzyme mechanism at atomic resolution. Science 294, 369–374 (2001).
Leung, D.W., Chen, E. & Goeddel, D.V. A method for random mutagenesis of a defined DNA segment using a modified polymerase chain reaction. Technique 1, 11–15 (1989).
Stemmer, W.P. DNA shuffling by random fragmentation and reassembly: in vitro recombination for molecular evolution. Proc. Natl. Acad. Sci. USA 91, 10747–10751 (1994).
Barbas, C.F. et al. Immune versus natural selection: antibody aldolases with enzymic rates but broader scope. Science 278, 2085–2092 (1997).
Esposito, G. et al. Structural and functional analysis of aldolase B mutants related to hereditary fructose intolerance. FEBS Lett. 531, 152–156 (2002).
Kipnis, Y. & Baker, D. Comparison of designed and randomly generated catalysts for simple chemical reactions. Protein Sci. 21, 1388–1395 (2012).
St-Jean, M., Blonski, C. & Sygusch, J. Charge stabilization and entropy reduction of central lysine residues in fructose-bisphosphate aldolase. Biochemistry 48, 4528–4537 (2009).
Wilson, I.A. & Stanfield, R.L. Antibody-antigen interactions: new structures and new conformational changes. Curr. Opin. Struct. Biol. 4, 857–867 (1994).
Cauerhff, A., Goldbaum, F.A. & Braden, B.C. Structural mechanism for affinity maturation of an anti-lysozyme antibody. Proc. Natl. Acad. Sci. USA 101, 3539–3544 (2004).
Ruscio, J.Z., Kohn, J.E., Ball, K.A. & Head-Gordon, T. The influence of protein dynamics on the success of computational enzyme design. J. Am. Chem. Soc. 131, 14111–14115 (2009).
Tokuriki, N. & Tawfik, D.S. Protein dynamism and evolvability. Science 324, 203–207 (2009).
Babbitt, P.C. & Gerlt, J.A. Understanding enzyme superfamilies. Chemistry as the fundamental determinant in the evolution of new catalytic activities. J. Biol. Chem. 272, 30591–30594 (1997).
Kuhlman, B. et al. Design of a novel globular protein fold with atomic-level accuracy. Science 302, 1364–1368 (2003).
Zanghellini, A. et al. New algorithms and an in silico benchmark for computational enzyme design. Protein Sci. 15, 2785–2794 (2006).
Neylon, C. Chemical and biochemical strategies for the randomization of protein encoding DNA sequences: library construction methods for directed evolution. Nucleic Acids Res. 32, 1448–1459 (2004).
Ho, S.N., Hunt, H.D., Horton, R.M., Pullen, J.K. & Pease, L.R. Site-directed mutagenesis by overlap extension using the polymerase chain-reaction. Gene 77, 51–59 (1989).
Kabsch, W. XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010).
McCoy, A.J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Hennig, M., Darimont, B.D., Jansonius, J.N. & Kirschner, K. The catalytic mechanism of indole-3-glycerol phosphate synthase: crystal structures of complexes of the enzyme from Sulfolobus solfataricus with substrate analogue, substrate, and product. J. Mol. Biol. 319, 757–766 (2002).
Adams, P.D. et al. Recent developments in the PHENIX software for automated crystallographic structure determination. J. Synchotron Radiat. 11, 53–55 (2004).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Karplus, P.A. & Diederichs, K. Linking crystallographic model and data quality. Science 336, 1030–1033 (2012).
List, B., Barbas, C.F. III & Lerner, R.A. Aldol sensors for the rapid generation of tunable fluorescence by antibody catalysis. Proc. Natl. Acad. Sci. USA 95, 15351–15355 (1998).
Turner, J.M., Bui, T., Lerner, R.A., Barbas, C.F. III & List, B. An efficient benchtop system for multigram-scale kinetic resolutions using aldolase antibodies. Chemistry 6, 2772–2774 (2000).
Acknowledgements
We thank T. Tomizaki, M. Müller, V. Olieric, G. Pompidor and A. Pauluhn at the Swiss Light Source for their outstanding support and all of the members of the Ban laboratory for suggestions and discussions. We are also grateful to E. Althoff (University of Washington) for providing the genes for RA95.0 and RA95.5 and sharing data before publication and to D. Gillingham and S. Tonazzi (ETH Zurich) for inhibitor synthesis and substrate resolution. The authors acknowledge support from the Swiss National Science Foundation (SNSF) (N.B. and D.H.), the National Center of Excellence in Research Structural Biology program of the SNSF (N.B.), the ETH Zurich (P.K., N.B. and D.H.), the Defense Advanced Research Projects Agency (D.B. and D.H.) and the Howard Hughes Medical Institute (D.B.). L.G. was funded by the Stipendienfonds der Schweizerischen Chemischen Industrie.
Author information
Authors and Affiliations
Contributions
D.H., N.B., D.B., P.K., L.G. and S.C. designed the experiments. L.G. and R.O. evolved and biochemically characterized the variants; S.C. crystallized the proteins and solved their structures. The manuscript and figures were prepared by L.G., S.C., P.K., N.B. and D.H.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results (PDF 3491 kb)
Rights and permissions
About this article
Cite this article
Giger, L., Caner, S., Obexer, R. et al. Evolution of a designed retro-aldolase leads to complete active site remodeling. Nat Chem Biol 9, 494–498 (2013). https://doi.org/10.1038/nchembio.1276
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.1276
This article is cited by
-
Bayesian optimization with evolutionary and structure-based regularization for directed protein evolution
Algorithms for Molecular Biology (2021)
-
Chance emergence of catalytic activity and promiscuity in a self-replicator
Nature Catalysis (2020)
-
Ensemble-based enzyme design can recapitulate the effects of laboratory directed evolution in silico
Nature Communications (2020)
-
Synthetic biology approaches to dissecting linear motor protein function: towards the design and synthesis of artificial autonomous protein walkers
Biophysical Reviews (2020)
-
The importance of catalytic promiscuity for enzyme design and evolution
Nature Reviews Chemistry (2019)