Abstract
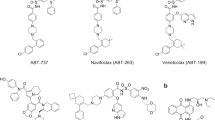
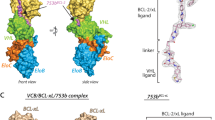
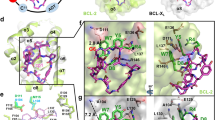
The prosurvival BCL-2 family protein BCL-XL is often overexpressed in solid tumors and renders malignant tumor cells resistant to anticancer therapeutics. Enhancing apoptotic responses by inhibiting BCL-XL will most likely have widespread utility in cancer treatment and, instead of inhibiting multiple prosurvival BCL-2 family members, a BCL-XL–selective inhibitor would be expected to minimize the toxicity to normal tissues. We describe the use of a high-throughput screen to discover a new series of small molecules targeting BCL-XL and their structure-guided development by medicinal chemistry. The optimized compound, WEHI-539 (7), has high affinity (subnanomolar) and selectivity for BCL-XL and potently kills cells by selectively antagonizing its prosurvival activity. WEHI-539 will be an invaluable tool for distinguishing the roles of BCL-XL from those of its prosurvival relatives, both in normal cells and notably in malignant tumor cells, many of which may prove to rely upon BCL-XL for their sustained growth.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Hotchkiss, R.S., Strasser, A., McDunn, J.E. & Swanson, P.E. Cell death. N. Engl. J. Med. 361, 1570–1583 (2009).
Strasser, A., Cory, S. & Adams, J.M. Deciphering the rules of programmed cell death to improve therapy of cancer and other diseases. EMBO J. 30, 3667–3683 (2011).
Glaser, S.P. et al. Anti-apoptotic Mcl-1 is essential for the development and sustained growth of acute myeloid leukemia. Genes Dev. 26, 120–125 (2012).
Lessene, G., Czabotar, P.E. & Colman, P.M. BCL-2 family antagonists for cancer therapy. Nat. Rev. Drug Discov. 7, 989–1000 (2008).
Wells, J.A. & McClendon, C.L. Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature 450, 1001–1009 (2007).
Mullard, A. Protein-protein interaction inhibitors get into the groove. Nat. Rev. Drug Discov. 11, 173–175 (2012).
Beroukhim, R. et al. The landscape of somatic copy-number alteration across human cancers. Nature 463, 899–905 (2010).
Amundson, S.A. et al. An informatics approach identifying markers of chemosensitivity in human cancer cell lines. Cancer Res. 60, 6101–6110 (2000).
Vaux, D.L., Cory, S. & Adams, J.M. Bcl-2 gene promotes haemopoietic cell survival and cooperates with c-myc to immortalize pre-B cells. Nature 335, 440–442 (1988).
Strasser, A., Harris, A.W., Bath, M.L. & Cory, S. Novel primitive lymphoid tumours induced in transgenic mice by cooperation between myc and bcl-2. Nature 348, 331–333 (1990).
Kelly, P.N. & Strasser, A. The role of Bcl-2 and its pro-survival relatives in tumourigenesis and cancer therapy. Cell Death Differ. 18, 1414–1424 (2011).
Lindsten, T. et al. The combined functions of proapoptotic Bcl-2 family members bak and bax are essential for normal development of multiple tissues. Mol. Cell 6, 1389–1399 (2000).
Leber, B., Lin, J. & Andrews, D.W. Still embedded together binding to membranes regulates Bcl-2 protein interactions. Oncogene 29, 5221–5230 (2010).
Leber, B., Lin, J. & Andrews, D.W. Embedded together: the life and death consequences of interaction of the Bcl-2 family with membranes. Apoptosis 12, 897–911 (2007).
Labi, V., Grespi, F., Baumgartner, F. & Villunger, A. Targeting the Bcl-2–regulated apoptosis pathway by BH3 mimetics: a breakthrough in anticancer therapy? Cell Death Differ. 15, 977–987 (2008).
Oltersdorf, T. et al. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature 435, 677–681 (2005).
Rudin, C.M. et al. Phase II study of single-agent navitoclax (ABT-263) and biomarker correlates in patients with relapsed small cell lung cancer. Clin. Cancer Res. 18, 3163–3169 (2012).
Gandhi, L. et al. Phase I study of navitoclax (ABT-263), a novel Bcl-2 family inhibitor, in patients with small-cell lung cancer and other solid tumors. J. Clin. Oncol. 29, 909–916 (2011).
Roberts, A.W. et al. Substantial susceptibility of chronic lymphocytic leukemia to BCL2 inhibition: results of a phase I study of navitoclax in patients with relapsed or refractory disease. J. Clin. Oncol. 30, 488–496 (2012).
Wilson, W.H. et al. Navitoclax, a targeted high-affinity inhibitor of BCL-2, in lymphoid malignancies: a phase 1 dose-escalation study of safety, pharmacokinetics, pharmacodynamics, and antitumour activity. Lancet Oncol. 11, 1149–1159 (2010).
Park, C.M. et al. Discovery of an orally bioavailable small molecule inhibitor of prosurvival B-cell lymphoma 2 proteins. J. Med. Chem. 51, 6902–6915 (2008).
Tse, C. et al. ABT-263: a potent and orally bioavailable Bcl-2 family inhibitor. Cancer Res. 68, 3421–3428 (2008).
Souers, A.J. et al. ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nat. Med. 19, 202–208 (2013).
Mason, K.D. et al. Programmed anuclear cell death delimits platelet life span. Cell 128, 1173–1186 (2007).
Zhang, H. et al. Bcl-2 family proteins are essential for platelet survival. Cell Death Differ. 14, 943–951 (2007).
Baell, J.B. & Holloway, G.A. New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J. Med. Chem. 53, 2719–2740 (2010).
Petros, A.M. et al. Rationale for Bcl-XL/Bad peptide complex formation from structure, mutagenesis, and biophysical studies. Protein Sci. 9, 2528–2534 (2000).
Petros, A.M., Olejniczak, E.T. & Fesik, S.W. Structural biology of the Bcl-2 family of proteins. Biochim. Biophys. Acta 1644, 83–94 (2004).
Lee, E.F. et al. Crystal structure of ABT-737 complexed with Bcl-XL: implications for selectivity of antagonists of the Bcl-2 family. Cell Death Differ. 14, 1711–1713 (2007).
Liu, X., Dai, S., Zhu, Y., Marrack, P. & Kappler, J. The structure of a Bcl-XL/Bim fragment complex: implications for Bim function. Immunity 19, 341–352 (2003).
Oberstein, A., Jeffrey, P.D. & Shi, Y. Crystal structure of the Bcl-XL–Beclin 1 peptide complex: Beclin 1 is a novel BH3-only protein. J. Biol. Chem. 282, 13123–13132 (2007).
Sattler, M. et al. Structure of Bcl-XL–Bak peptide complex: recognition between regulators of apoptosis. Science 275, 983–986 (1997).
Lee, E.F. et al. Conformational changes in Bcl-2 pro-survival proteins determine their capacity to bind ligands. J. Biol. Chem. 284, 30508–30517 (2009).
Wendt, M.D. et al. Discovery and structure-activity relationship of antagonists of B-cell lymphoma 2 family proteins with chemopotentiation activity in vitro and in vivo. J. Med. Chem. 49, 1165–1181 (2006).
Chen, L. et al. Differential targeting of prosurvival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol. Cell 17, 393–403 (2005).
Czabotar, P.E. & Lessene, G. Bcl-2 family proteins as therapeutic targets. Curr. Pharm. Des. 16, 3132–3148 (2010).
van Delft, M.F. et al. The BH3 mimetic ABT-737 targets selective Bcl-2 proteins and efficiently induces apoptosis via Bak/Bax if Mcl-1 is neutralized. Cancer Cell 10, 389–399 (2006).
van Delft, M.F. et al. The BH3 mimetic ABT-737 targets selective Bcl-2 proteins and efficiently induces apoptosis via Bak/Bax if Mcl-1 is neutralized. Cancer Cell 10, 389–399 (2006).
Willis, S.N. et al. Proapoptotic Bak is sequestered by Mcl-1 and Bcl-XL, but not Bcl-2, until displaced by BH3-only proteins. Genes Dev. 19, 1294–1305 (2005).
Lee, E.F. et al. A novel BH3 ligand that selectively targets Mcl-1 reveals that apoptosis can proceed without Mcl-1 degradation. J. Cell Biol. 180, 341–355 (2008).
Mérino, D. et al. Bcl-2, Bcl-XL, and Bcl-W are not equivalent targets of ABT-737 and navitoclax (ABT-263) in lymphoid and leukemic cells. Blood 119, 5807–5816 (2012).
Josefsson, E.C. et al. Megakaryocytes possess a functional intrinsic apoptosis pathway that must be restrained to survive and produce platelets. J. Exp. Med. 208, 2017–2031 (2011).
Gleeson, M.P. Generation of a set of simple, interpretable ADMET rules of thumb. J. Med. Chem. 51, 817–834 (2008).
Shoemaker, A.R. et al. A small-molecule inhibitor of Bcl-XL potentiates the activity of cytotoxic drugs in vitro and in vivo. Cancer Res. 66, 8731–8739 (2006).
Czabotar, P.E. et al. Structural insights into the degradation of Mcl-1 induced by BH3 domains. Proc. Natl. Acad. Sci. USA 104, 6217–6222 (2007).
Willis, S.N. et al. Apoptosis initiated when BH3 ligands engage multiple Bcl-2 homologs, not Bax or Bak. Science 315, 856–859 (2007).
Fletcher, J.I. et al. Apoptosis is triggered when prosurvival Bcl-2 proteins cannot restrain Bax. Proc. Natl. Acad. Sci. USA 105, 18081–18087 (2008).
Smits, C., Czabotar, P.E., Hinds, M.G. & Day, C.L. Structural plasticity underpins promiscuous binding of the prosurvival protein A1. Structure 16, 818–829 (2008).
Muchmore, S.W. et al. X-ray and NMR structure of human Bcl-XL, an inhibitor of programmed cell death. Nature 381, 335–341 (1996).
Kvansakul, M. et al. Vaccinia virus anti-apoptotic F1L is a novel Bcl-2–like domain-swapped dimer that binds a highly selective subset of BH3-containing death ligands. Cell Death Differ. 15, 1564–1571 (2008).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276, 307–326 (1997).
Kabsch, W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr. D Biol. Crystallogr. 66, 133–144 (2010).
McCoy, A.J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Murshudov, G.N., Vagin, A.A. & Dodson, E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 (1997).
Adams, P.D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Acknowledgements
The authors thank J. Blyth, D. Buczek, A. Georgiou, H. Ierino, C. Rye, G. Siciliano, G. Thompson and A. Wardak for outstanding technical assistance; S. Cory (Walter and Eliza Hall Institute of Medical Research (WEHI)), L. Chen (WEHI), V. Dixit (Genentech (GNE)), M. Hijnen (GE Healthcare), S. Hymowitz (GNE), D. Segal (WEHI), V. Tsui (GNE), M.F. van Delft (WEHI), A.H. Wei (WEHI) and I. Wertz (GNE) for discussions and suggestions; P. Bouillet and A. Strasser (both from WEHI) for mice; W.D. Fairlie and E. Lee (both from WEHI) for expression constructs; W. Welch (University of California–San Francisco) for anti-HSP70; P. Pilling, V. Streltsov (both from Commonwealth Scientific and Industrial Research Organisation, Australia) and staff at the photon Factory BL6 for their help with collecting data for 2; and AbbVie for providing ABT-737. This work was supported by fellowships and grants from the Australian Research Council (fellowship to P.E.C.), the National Health and Medical Research Council (NHMRC) (fellowships to J.M.A., J.B.B., P.M.C. and D.C.S.H.; development grant 305536 and program grants 257502, 461221 and 1016701), the Leukemia and Lymphoma Society (specialized center of research grant nos. 7015 and 7413), the Cancer Council of Victoria (fellowship to P.M.C.; grant-in-aid 461239) and the Australian Cancer Research Foundation. Infrastructure support from the NHMRC Independent Research Institutes Infrastructure Support Scheme grant no. 361646 and a Victorian State Government OIS grant are gratefully acknowledged.
Author information
Authors and Affiliations
Contributions
G.L. devised synthetic strategies, synthesized compounds, supervised the chemistry team and wrote the manuscript. P.E.C. performed X-ray crystallization and solved and analyzed structures. B.E.S., K.Z., W.J.A.K., S.K. and K.D. designed and synthesized compounds. J.M.A. conceived the study and analyzed data and results. J.B.B., K.D., J.A.F. and K.G.W. designed synthetic strategies and oversaw chemistry efforts. P.M.C. conceived the study and designed and oversaw structural studies. K.N.L. performed biological experiments. P.G. and B.J.S. performed computational modeling. W.J.F. and J.A.F. designed studies and data analysis. R.M.M., J.P.P. and I.P.S. performed the high-throughput chemical screening campaign. H.Y. performed Biacore experiments. D.C.S.H. conceived the study, designed and oversaw biological experiments and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
G.L., P.E.C., B.E.S., K.N.L., J.M.A., J.B.B., P.M.C., W.J.A.K., S.K., R.M.M., J.P.P., B.J.S., I.P.S., H.Y., D.C.S.H. and K.G.W. are or were employees of WEHI, which received commercial income and research funding from Genentech, Inc. for part of this work. W.J.F., J.A.F., K.D., P.G. and K.Z. are employees of Genentech, Inc., a member of the Roche group, and are stockholders of Roche Holding.
Supplementary information
Supplementary Text and Figures
Supplementary Results (PDF 10321 kb)
Supplementary Note 1
Supplementary Note 1 (PDF 1219 kb)
Supplementary Note 2
Supplementary Note 2 (PDF 527 kb)
Rights and permissions
About this article
Cite this article
Lessene, G., Czabotar, P., Sleebs, B. et al. Structure-guided design of a selective BCL-XL inhibitor. Nat Chem Biol 9, 390–397 (2013). https://doi.org/10.1038/nchembio.1246
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.1246
This article is cited by
-
Mechanisms of BCL-2 family proteins in mitochondrial apoptosis
Nature Reviews Molecular Cell Biology (2023)
-
BCL-2 protein family: attractive targets for cancer therapy
Apoptosis (2023)
-
Synthesis and in vitro anticancer potential of new thiazole-containing derivatives of rhodanine
Chemistry of Heterocyclic Compounds (2023)
-
Targeting the apoptosis pathway to treat tumours of the paediatric nervous system
Cell Death & Disease (2022)
-
Noxa and Mcl-1 expression influence the sensitivity to BH3-mimetics that target Bcl-xL in patient-derived glioma stem cells
Scientific Reports (2022)