Abstract
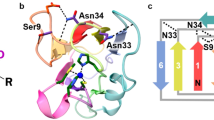
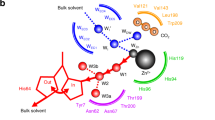
Many proteins contain copper in a range of coordination environments, where it has various biological roles, such as transferring electrons or activating dioxygen. These copper sites can be classified by their function or spectroscopic properties. Those with a single copper atom are either type 1, with an intense absorption band near 600 nm, or type 2, with weak absorption in the visible region. We have built a novel copper(ii) binding site within structurally modified Pseudomonas aeruginosa azurins that does not resemble either existing type, which we therefore call ‘type zero’. X-ray crystallographic analysis shows that these sites adopt distorted tetrahedral geometries, with an unusually short Cu–O (G45 carbonyl) bond. Relatively weak absorption near 800 nm and narrow parallel hyperfine splittings in electron paramagnetic resonance spectra are the spectroscopic signatures of type zero copper. Cyclic voltammetric experiments demonstrate that the electron transfer reactivities of type-zero azurins are enhanced relative to that of the corresponding type 2 (C112D) protein.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Gray, H. B. Biological inorganic chemistry at the beginning of the 21st century. Proc. Natl Acad. Sci. USA 100, 3563–3568 (2003).
Gradinaru, C., Crane, B. R., Abrahamsson, M. L. & Gray, H. B. Electron transfer in metalloproteins (blue copper azurin). Biophys. J. 86, 473A (2004).
Roberts, S. A. et al. Crystal structure and electron transfer kinetics of CueO, a multicopper oxidase required for copper homeostasis in Escherichia coli. Proc. Natl Acad. Sci. USA 99, 2766–2771 (2002).
Palmer, A. E. et al. Spectroscopic characterization and O-2 reactivity of the trinuclear Cu cluster of mutants of the multicopper oxidase Fet3p. Biochemistry 41, 6438–6448 (2002).
Solomon, E. I., Szilagyi, R. K., DeBeer George, S. & Basumallick, L. Electronic structures of metal sites in proteins and models: Contributions to function in blue copper proteins. Chem. Rev. 104, 419–458 (2004).
Solomon, E. I. Spectroscopic methods in bioinorganic chemistry: Blue to green to red copper sites. Inorg. Chem. 45, 8012–8025 (2006).
Lee, S. K. et al. Nature of the intermediate formed in the reduction of O2 to H2O at the trinuclear copper cluster active site in native laccase. J. Am. Chem. Soc. 124, 6180–6193 (2002).
Hay, M., Richards, J. H. & Lu, Y. Construction and characterization of an azurin analog for the purple copper site in cytochrome c oxidase. Proc. Natl Acad. Sci. USA 93, 461–464 (1996).
Mizoguchi, T. J., Di Bilio, A. J., Gray, H. B. & Richards, J. H. Blue to type 2 binding. Copper(ii) and cobalt (ii) derivatives of a Cys112Asp mutant of Pseudomonas aeruginosa azurin. J. Am. Chem. Soc. 114, 10076–10078 (1992).
Mizoguchi, T. J. Probing the role of the active site cysteine of azurin by site-directed mutagenesis. PhD thesis, California Institute of Technology (1996).
Lancaster, K. M., Yokoyama, K., Richards, J. H., Winkler, J. R. & Gray, H. B. High-potential C112D/M121X (X = M, E, H, L) Pseudomonas aeruginosa azurins. Inorg. Chem. 48, 1278–1280 (2009).
Murshudov, G. N., Vagin, A. A. & Dodson, E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53, 240–255 (1997).
Vaguine, A. A., Richelle, J. & Wodak, S. J. SFCHECK: a unified set of procedures for evaluating the quality of macromolecular structure-factor data and their agreement with the atomic model. Acta Crystallogr. D 55, 191–205 (1999).
Engman, K. C., Sandberg, A., Leckner, J. & Karlsson, B. G. Probing the influence on folding behavior of structurally conserved core residues in P-aeruginosa apo-azurin. Protein Sci. 13, 2706–2715 (2004).
Faham, S. et al. Role of the active-site cysteine of Pseudomonas aeruginosa azurin. Crystal structure analysis of the CuII(Cys112Asp) protein. J. Biol. Inorg. Chem. 2, 464–469 (1997).
Gray, H. B., Malmstrom, B. G. & Williams, R. J. P. Copper coordination in blue proteins. J. Biol. Inorg. Chem. 5, 551–559 (2000).
DeBeer, S. et al. X-ray absorption spectra of the oxidized and reduced forms of C112D azurin from Pseudomonas aeruginosa. Inorg. Chem. 38, 433–438 (1999).
Hitchman, M. A., Olson, C. D. & Belford, R. L. Behavior of in-plane g-tensor in low-symmetry d1 and d9 systems with application to copper and vanadyl chelates. J. Chem. Phys. 50, 1195–1203 (1969).
Gewirth, A. A., Cohen, S. L., Schugar, H. J. & Solomon, E. I. Spectroscopic and theoretical studies of the unusual electron-paramagnetic-resonance parameters of distorted tetrahedral cupric sites - correlations to X-ray spectral features of core levels. Inorg. Chem. 26, 1133–1146 (1987).
Mabbs, F. E. & Machin, D. J. Magnetism and Transition Metal Complexes (Dover Publications, 2008).
Stevens, K. W. H. On The magnetic properties of covalent XY6 complexes. Proc. R. Soc. Lond. A 219, 542–555 (1953).
Griffiths, J. H. E. & Owen, J. Complex hyperfine structures in microwave spectra of covalent iridium compounds. Proc. R. Soc. Lond. A 226, 96–111 (1954).
Fujita, K. et al. Mimicking protein-protein electron transfer: Voltammetry of Pseudomonas aeruginosa azurin and the Thermus thermophilus Cu-A domain at omega-derivatized self-assembled-monolayer gold electrodes. J. Am. Chem. Soc. 126, 13954–13961 (2004).
Ghosh, S., Xie, X., Dey, A., Sun, Y., Scholes, C. P. & Solomon, E. I. Thermodynamic equilibrium between blue and green copper sites and the role of the protein in controlling function. Proc. Natl Acad. Sci. USA 106, 4969–4974 (2009).
Miura, Y. et al. Direct electrochemistry of CueO and its mutants at residues to and near Type I Cu for oxygen-reducing biocathode. Fuel Cells 9, 70–78 (2009).
Tsujimura, S., Miura, Y. & Kano, K. CueO-immobilized porous carbon electrode exhibiting improved performance of electrochemical reduction of dioxygen to water. Electrochimica Acta 53, 5716–5720 (2008).
Miura, Y. et al. Bioelectrocatalytic reduction of O-2 catalyzed by CueO from Escherichia coli adsorbed on a highly oriented pyrolytic graphite electrode. Chem. Lett. 36, 132–133 (2007).
Blanford, C. F., Foster, C. E., Heath, R. S. & Armstrong, F. A. Efficient electrocatalytic oxygen reduction by the ‘blue’ copper oxidase, laccase, directly attached to chemically modified carbons. Faraday Discuss. 140, 319–335 (2008).
Leslie, A. G. W. Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography 26 (1992).
CCP4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994).
Cohen, S. X. et al. ARP/wARP and molecular replacement: the next generation. Acta Crystallogr. D 64, 49–60 (2008).
Golombek, A. P. & Hendrich, M. P. Quantitative analysis of dinuclear manganese(ii) EPR spectra. J. Magn. Reson. 165, 33–48 (2003).
George, G. N. EXAFSPAK (Stanford Synchrotron Radiation Laboratory, Stanford Linear Accelerator Center, Stanford University).
Tenderhold, A. PySpline (Stanford Synchrotron Radiation Laboratory, Stanford Linear Accelerator Center, Stanford University).
DeLeon, J. M., Rehr, J. J., Zabinsky, S. I. & Albers, R. C. Abinitio curved-wave X-ray-absorption fine-structure. Phys. Rev. B 44, 4146–4156 (1991).
Rehr, J. J., DeLeon, J. M., Zabinsky, S. I. & Albers, R. C. Theoretical X-ray absorption fine-structure standards. J. Am. Chem. Soc. 113, 5135–5140 (1991).
Yokoyama, K. et al. Electron tunneling through Pseudomonas aeruginosa azurins on SAM gold electrodes. Inorg. Chim. Acta 361, 1095–1099 (2008).
Laviron, E. General expression of the linear potential sweep voltammogram in the case of diffusionless electrochemical systems. J. Electroanal. Chem. 101, 19–28 (1979).
Pascher, T., Karlsson, B. G., Nordling, M., Malmstrom, B. G. & Vanngard, T. Reduction potentials and their pH dependence in site-directed-mutant forms of azurin from Pseudomonas aeruginosa. Eur. J. Biochem. 212, 289–296 (1993).
Acknowledgements
We thank B. Brunschwig for assistance with Fourier transform infrared spectroscopy, Z. Gates and L. Thomas for assistance with X-ray diffraction data collection, and M. Day and J. Kaiser for discussions of crystal structural analyses. We thank E. Solomon for helpful comments on electronic structural formulations, and Y. Sheng for assistance with protein expression and purification. Stanford Synchrotron Radiation Lightsource operations are funded by DOE(BES). The Structural Molecular Biology program is supported by NIH (NCRR BMTP) (Grant Number 5 P41 RR001209)N and DOE(BER). This work was supported by NIH DK019038(HBG), Stanford GCEP, and NSF CHE-0802907. The Caltech Molecular Observatory is supported by the Gordon and Betty Moore Foundation.
Author information
Authors and Affiliations
Contributions
K.M.L. and H.B.G. conceived and designed the experiments; K.M.L., S.D.G., and K.Y. performed the experiments; K.M.L., S.D.G., K.Y., and H.B.G. analysed the data, K.M.L., S.D.G., J.H.R., and H.B.G. co-wrote the paper.
Corresponding authors
Supplementary information
Supplementary information
Supplementary information (PDF 2637 kb)
Rights and permissions
About this article
Cite this article
Lancaster, K., George, S., Yokoyama, K. et al. Type-zero copper proteins. Nature Chem 1, 711–715 (2009). https://doi.org/10.1038/nchem.412
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchem.412
This article is cited by
-
Transferring the entatic-state principle to copper photochemistry
Nature Chemistry (2018)
-
Reversible S-nitrosylation in an engineered azurin
Nature Chemistry (2016)
-
Zeroing in on a new copper site
Nature Chemistry (2009)