Abstract
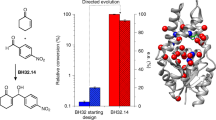
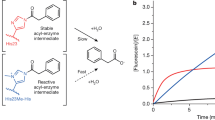
Random mutagenesis has the potential to optimize the efficiency and selectivity of protein catalysts without requiring detailed knowledge of protein structure; however, introducing synthetic metal cofactors complicates the expression and screening of enzyme libraries, and activity arising from free cofactor must be eliminated. Here we report an efficient platform to create and screen libraries of artificial metalloenzymes (ArMs) via random mutagenesis, which we use to evolve highly selective dirhodium cyclopropanases. Error-prone PCR and combinatorial codon mutagenesis enabled multiplexed analysis of random mutations, including at sites distal to the putative ArM active site that are difficult to identify using targeted mutagenesis approaches. Variants that exhibited significantly improved selectivity for each of the cyclopropane product enantiomers were identified, and higher activity than previously reported ArM cyclopropanases obtained via targeted mutagenesis was also observed. This improved selectivity carried over to other dirhodium-catalysed transformations, including N–H, S–H and Si–H insertion, demonstrating that ArMs evolved for one reaction can serve as starting points to evolve catalysts for others.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
References
Mahatthananchai, J., Dumas, A. M. & Bode, J. W. Catalytic selective synthesis. Angew. Chem. Int. Ed. 51, 10954–10990 (2012).
Bornscheuer, U. T. et al. Engineering the third wave of biocatalysis. Nature 485, 185–194 (2012).
Romero, P. A. & Arnold, F. H. Exploring protein fitness landscapes by directed evolution. Nat. Rev. Mol. Cell Biol. 10, 866–876 (2009).
Lu, Y., Berry, S. & Pfister, T. Engineering novel metalloproteins: design of metal-binding sites into native protein scaffolds. Chem. Rev. 101, 3047–3080 (2001).
Gerlt, J. A. & Babbitt, P. C. Enzyme (re)design: lessons from natural evolution and computation. Curr. Opin. Chem. Biol. 13, 10–18 (2009).
Chen, K. & Arnold, F. H. Tuning the activity of an enzyme for unusual environments: sequential random mutagenesis of subtilisin E for catalysis in dimethylformamide. Proc. Natl Acad. Sci. USA 90, 5618–5622 (1993).
Stemmer, W. P. C. Rapid evolution of a protein in vitro by DNA shuffling. Nature 370, 389–391 (1994).
Lutz, S. Beyond directed evolution–semi-rational protein engineering and design. Curr. Opin. Biotech. 21, 734–743 (2010).
Lewis, J. C. Artificial metalloenzymes and metallopeptide catalysts for organic synthesis. ACS Catal. 3, 2954–2975 (2013).
Kazlauskas, R. J. & Bornscheuer, U. T. Finding better protein engineering strategies. Nat. Chem. Biol. 5, 526–529 (2009).
Reetz, M. T., Peyralans, J. J. P., Maichele, A., Fu, Y. & Maywald, M. Directed evolution of hybrid enzymes: evolving enantioselectivity of an achiral Rh-complex anchored to a protein. Chem. Commun. 2016, 4318–4320 (2006).
Jeschek, M. et al. Directed evolution of artificial metalloenzymes for in vivo metathesis. Nature 537, 661–665 (2016).
Song, W. J. & Tezcan, F. A. A designed supramolecular protein assembly with in vivo enzymatic activity. Science 346, 1525–1528 (2014).
Key, H. M., Dydio, P., Clark, D. S. & Hartwig, J. F. Abiological catalysis by artificial haem proteins containing noble metals in place of iron. Nature 534, 534–537 (2016).
Hyster, T. K. Genetic optimization of metalloenzymes: enhancing enzymes for non-natural reactions. Angew. Chem. Int. Ed. 55, 7344–7357 (2016).
Sreenilayam, G., Moore, E. J., Steck, V. & Fasan, R. Metal substitution modulates the reactivity and extends the reaction scope of myoglobin carbene transfer catalysts. Adv. Synth. Catal. 359, 2076–2089 (2017).
Sreenilayam, G., Moore, E. J., Steck, V. & Fasan, R. Stereoselective olefin cyclopropanation under aerobic conditions with an artificial enzyme incorporating an iron-chlorin e6 cofactor. ACS Catal. 7, 7629–7633 (2017).
Srivastava, P., Yang, H., Ellis-Guardiola, K. & Lewis, J. C. Engineering a dirhodium artificial metalloenzyme for selective olefin cyclopropanation. Nat. Commun. 6, 7789 (2015).
Juhász, T., Szeltner, Z. & Polgár, L. Properties of the prolyl oligopeptidase homologue from Pyrococcus furiosus. FEBS Lett. 580, 3493–3497 (2006).
Polgár, L. The prolyl oligopeptidase family. Cell. Mol. Life Sci. 59, 349–362 (2002).
Yang, H., Srivastava, P., Zhang, C. & Lewis, J. C. A general method for artificial metalloenzyme formation through strain-promoted azide–alkyne cycloaddition. ChemBioChem 15, 223–227 (2014).
Harris, M. N., Madura, J. D., Ming, L.-J. & Harwood, V. J. Kinetic and mechanistic studies of prolyl oligopeptidase from the hyperthermophile Pyrococcus furiosus. J. Biol. Chem. 276, 19310–19317 (2001).
Wilson, Y. M., Dürrenberger, M., Nogueira, E. S. & Ward, T. R. Neutralizing the detrimental effect of glutathione on precious metal catalysts. J. Am. Chem. Soc. 136, 8928–8932 (2014).
Reetz, M. T. et al. A robust protein host for anchoring chelating ligands and organocatalysts. ChemBioChem 9, 552–564 (2008).
Drummond, D., Iverson, B., Georgiou, G. & Arnold, F. H. Why high-error-rate random mutagenesis libraries are enriched in functional and improved proteins. J. Mol. Biol. 350, 806–816 (2005).
Dąbrowski, S. & Kur, J. Cloning and expression in Escherichia coli of the recombinant his-tagged DNA polymerases from Pyrococcus furiosus and Pyrococcus woesei. Protein Expr. Purif. 14, 131–138 (1998).
Punna, S., Kaltgrad, E. & Finn, M. G. ‘Clickable’ agarose for affinity chromatography. Bioconj. Chem. 16, 1536–1541 (2005).
Davies, R. R. et al. Artificial metalloenzymes based on protein cavities: exploring the effect of altering the metal ligand attachment position by site directed mutagenesis. Bioorg. Med. Chem. Lett. 9, 79–84 (1999).
Filice, M. et al. Synthesis of a heterogeneous artificial metallolipase with chimeric catalytic activity. Chem. Commun. 51, 9324–9327 (2015).
Ball, Z. T. Designing enzyme-like catalysts: a rhodium(II) metallopeptide case study. Acc. Chem. Res. 46, 560–570 (2013).
Renata, H. et al. Identification of mechanism-based inactivation in P450-catalyzed cyclopropanation facilitates engineering of improved enzymes. J. Am. Chem. Soc. 138, 12527–12533 (2016).
Popp, B. V. & Ball, Z. T. Proximity-driven metallopeptide catalysis: remarkable side-chain scope enables modification of the Fos bZip domain. Chem. Sci. 2, 690–695 (2011).
Lewis, J. C. & Arnold, F. H. Catalysts on demand: selective oxidations by laboratory-evolved cytochrome P450 BM3. CHIMIA 63, 309–312 (2009).
Lu, Y., Yeung, N., Sieracki, N. & Marshall, N. M. Design of functional metalloproteins. Nature 460, 855–862 (2009).
Tokuriki, N. & Tawfik, D. S. Protein dynamism and evolvability. Science 324, 203–207 (2009).
Kaushik, S., Etchebest, C. & Sowdhamini, R. Decoding the structural events in substrate-gating mechanism of eukaryotic prolyl oligopeptidase using normal mode analysis and molecular dynamics simulations. Proteins 82, 1428–1443 (2014).
Acknowledgements
This work was supported by, or in part by, the US Army Research Laboratory and the US Army Research Office under contract/grant nos. W911NF-14-1-0334 and 66796-LS-RIP (to J.C.L.), the National Science Foundation (NSF) under CAREER Award CHE-1351991 (to J.C.L.), The David and Lucile Packard Foundation (to J.C.L.), the National Institutes of Health (NIH) National Cancer Institute (R00CA175399 to R.E.M.), the Damon Runyon Cancer Research Foundation (DFS-08-14 to R.E.M.) and the NSF under the Center for Chemical Innovation Center for Selective C–H Functionalization (CHE-1700982, to J.C.L.). K.E.G. and D.M.U. were funded by an NIH Chemistry and Biology Interface Training Grant (T32 GM008720) and G.L. was supported by the Kwanjeong Educational Foundation. MS data were acquired on instruments purchased using an NSF instrumentation grant (CHE-1048528).
Author information
Authors and Affiliations
Contributions
H.Y. and P.S. developed the soluble ArM evolution procedure. C.Z. optimized the soluble ArM evolution procedure. A.M.S. and D.M.U. optimized the soluble ArM evolution procedure and collected conversion and selectivity data for all evolved ArMs. H.J.P. optimized and executed the immobilized ArM evolution procedure. K.B. and Y.G. cloned POP amber mutants and the combinatorial codon mutagenesis library. K.E.-G., G.L. and R.E.M. conducted and analysed the LC-MS/MS experiments. J.C.L. devised the experiments and procedures, designed the ArM variants and libraries, analysed data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information
Supplementary information
Rights and permissions
About this article
Cite this article
Yang, H., Swartz, A., Park, H. et al. Evolving artificial metalloenzymes via random mutagenesis. Nature Chem 10, 318–324 (2018). https://doi.org/10.1038/nchem.2927
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchem.2927
This article is cited by
-
Directed evolution of a wax ester synthase for production of fatty acid ethyl esters in Saccharomyces cerevisiae
Applied Microbiology and Biotechnology (2023)
-
Engineering and emerging applications of artificial metalloenzymes with whole cells
Nature Catalysis (2021)
-
High performance crystalline nanocellulose using an ancestral endoglucanase
Communications Materials (2020)
-
Expanding the enzyme universe with genetically encoded unnatural amino acids
Nature Catalysis (2020)
-
Engineering new catalytic activities in enzymes
Nature Catalysis (2020)