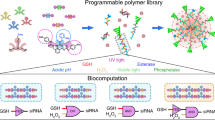
Abstract
The successful transport of drug- and cell-based therapeutics to diseased sites represents a major barrier in the development of clinical therapies. Targeted delivery can be mediated through degradable biomaterial vehicles that utilize disease biomarkers to trigger payload release. Here, we report a modular chemical framework for imparting hydrogels with precise degradative responsiveness by using multiple environmental cues to trigger reactions that operate user-programmable Boolean logic. By specifying the molecular architecture and connectivity of orthogonal stimuli-labile moieties within material cross-linkers, we show selective control over gel dissolution and therapeutic delivery. To illustrate the versatility of this methodology, we synthesized 17 distinct stimuli-responsive materials that collectively yielded all possible YES/OR/AND logic outputs from input combinations involving enzyme, reductant and light. Using these hydrogels we demonstrate the first sequential and environmentally stimulated release of multiple cell lines in well-defined combinations from a material. We expect these platforms will find utility in several diverse fields including drug delivery, diagnostics and regenerative medicine.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Burdick, J. A. & Murphy, W. L. Moving from static to dynamic complexity in hydrogel design. Nat. Commun. 3, 1269 (2012).
Hoffman, A. S. Stimuli-responsive polymers: biomedical applications and challenges for clinical translation. Adv. Drug Deliv. Rev. 65, 10–16 (2013).
Knipe, J. M. & Peppas, N. A. Multi-responsive hydrogels for drug delivery and tissue engineering applications. Regen. Biomater. 1, 57–65 (2014).
Mura, S., Nicolas, J. & Couvreur, P. Stimuli-responsive nanocarriers for drug delivery. Nat. Mater. 12, 991–1003 (2013).
O'Neill, H. S. et al. Biomaterial-enhanced cell and drug delivery: lessons learned in the cardiac field and future perspectives. Adv. Mater. 28, 5648–5661 (2016).
Tibbitt, M. W., Rodell, C. B., Burdick, J. A. & Anseth, K. S. Progress in material design for biomedical applications. Proc. Natl Acad. Sci. USA 112, 14444–14451 (2015).
Evans, A. C., Thadani, N. N. & Suh, J. Biocomputing nanoplatforms as therapeutics and diagnostics. J. Control. Rel. 240, 387–393 (2016).
Lu, Y., Aimetti, A. A., Langer, R. & Gu, Z. Bioresponsive materials. Nat. Rev. Mater. 2, 16075 (2016).
DeForest, C. A. & Anseth, K. S. Advances in bioactive hydrogels to probe and direct cell fate. Annu. Rev. Chem. Biomol. Eng. 3, 421–444 (2012).
Li, J. & Mooney, D. J. Designing hydrogels for controlled drug delivery. Nat. Rev. Mater. 1, 16071 (2016).
McCawley, L. J. & Matrisian, L. M. Matrix metalloproteinases: multifunctional contributors to tumor progression. Mol. Med. Today 6, 149–156 (2000).
Chen, X. et al. Dual bioresponsive mesoporous silica nanocarrier as an ‘AND’ logic gate for targeted drug delivery cancer cells. Adv. Funct. Mater. 24, 6999–7006 (2014).
Choh, S., Cross, D. & Wang, C. Facile synthesis and characterization of disulfide-cross-linked hyaluronic acid hydrogels for protein delivery and cell encapsulation. Biomacromolecules 12, 1126–1136 (2011).
Douglas, S. M., Bachelet, I. & Church, G. M. A logic-gated nanorobot for targeted transport of molecular payloads. Science 335, 831–834 (2012).
Ikeda, M. et al. Installing logic-gate responses to a variety of biological substances in supramolecular hydrogel-enzyme hybrids. Nat. Chem. 6, 511–518 (2014).
Komatsu, H. et al. Supramolecular hydrogel exhibiting four basic logic gate functions to fine-tune substance release. J. Am. Chem. Soc. 131, 5580–5585 (2009).
Liu, G., Ji, W. & Feng, C. Installing logic gates to multiresponsive supramolecular hydrogel co-assembled from phenylalanine amphiphile and bis(pyridinyl) derivative. Langmuir 31, 7122–7128 (2015).
Motornov, M. et al. ‘Chemical transformers’ from nanoparticle ensembles operated with logic. Nano Lett. 8, 2993–2997 (2008).
Kharkar, P. M., Kiick, K. L. & Kloxin, A. M. Design of thiol- and light-sensitive degradable hydrogels using Michael-type additon reactions. Polym. Chem. 6, 5565–5574 (2015).
de Garcia Lux, C. et al. Short soluble coumarin crosslinkers for light-controlled release of cells and proteins from hydrogels. Biomacromolecules 16, 3286–3296 (2015).
Roche, E. T. et al. Comparison of biomaterial delivery vehicles for improving acute retention of stem cells in the infarcted heart. Biomaterials 35, 6850–6858 (2014).
Steinhilber, D. et al. A microgel construction kit for bioorthogonal encapsulation and pH-controlled release of living cells. Angew. Chem. Int. Ed. 52, 13538–13543 (2013).
Griffin, D. R. & Kasko, A. M. Photodegradable macromers and hydrogels for live cell encapsulation and release. J. Am. Chem. Soc. 134, 13103–13107 (2012).
Nagase, H. & Fields, G. B. Human matrix metalloproteinase specificity studies using collagen sequence-based synthetic peptides. Pept. Sci. 40, 399–416 (1996).
Kloxin, A. M., Tibbitt, M. W. & Anseth, K. S. Synthesis of photodegradable hydrogels as dynamically tunable cell culture platforms. Nat. Protoc. 5, 1867–1887 (2010).
Agard, N. J., Prescher, J. A. & Bertozzi, C. R. A strain-promoted [3+2] azide–alkyne cycloaddition for covalent modification of biomolecules in living systems. J. Am. Chem. Soc. 126, 15046–15047 (2004).
DeForest, C. A. & Anseth, K. S. Cytocompatible click-based hydrogels with dynamically tunable properties through orthogonal photoconjugation and photocleavage reactions. Nat. Chem. 3, 925–931 (2011).
DeForest, C. A., Polizzotti, B. D. & Anseth, K. S. Sequential click reactions for synthesizing and patterning three-dimensional cell microenvironments. Nat. Mater. 8, 659–664 (2009).
DeForest, C. A. & Tirrell, D. A. A photoreversible protein-patterning approach for guiding stem cell fate in three-dimensional gels. Nat. Mater. 14, 523–531 (2015).
Madl, C. M., Katz, L. M. & Heilshorn, S. C. Bio-orthogonally crosslinked, engineered protein hydrogels with tunable mechanics and biochemistry for cell encapsulation. Adv. Funct. Mater. 26, 3612–3620 (2016).
Jiang, Y., Chen, J., Deng, C., Suuronen, E. J. & Zhong, Z. Click hydrogels, microgels and nanogels: emerging platforms for drug delivery and tissue engineering. Biomaterials 35, 4969–4985 (2014).
Das, R. K., Gocheva, V., Hammink, R., Zouani, O. F. & Rowan, A. E. Stress-stiffening-mediated stem-cell commitment switch in soft responsive hydrogels. Nat. Mater. 15, 318–325 (2016).
Hiemenz, P. C. & Lodge, T. P. Polymer Chemistry (CRC Press, 2007).
Flynn, B. P. et al. Mechanical strain stabilizes reconstituted collagen fibrils against enzymatic degradation by mammalian collagenase matrix metalloproteinase 8 (MMP-8). PLoS ONE 5, e12337 (2010).
Huo, M., Yuan, J. & Wei, Y. Redox-responsive polymers for drug delivery: from molecular design to applications. Polym. Chem. 5, 1519–1528 (2014).
Zhu, C., Ninh, C. & Bettinger, C. J. Photoreconfigurable polymers for biomedical applications: chemistry and macromolecular engineering. Biomacromolecules 15, 3474–3494 (2014).
Uhrich, K. E., Cannizzaro, S. M., Langer, R. S. & Shakesheff, K. M. Polymeric systems for controlled drug release. Chem. Rev. 99, 3181–3198 (1999).
Arakawa, C. K., Badeau, B. A., Zheng, Y. & DeForest, C. A. Multicellular vascularized engineered tissues through user-programmable biomaterial photodegradation. Adv. Mater. 29, 1703156 (2017).
Khetan, S. & Burdick, J. A. Patterning network structure to spatially control cellular remodeling and stem cell fate within 3-dimensional hydrogels. Biomaterials 31, 8228–8234 (2010).
Khetan, S. et al. Degradation-mediated cellular traction directs stem cell fate in covalently crosslinked three-dimensional hydrogels. Nat. Mater. 12, 458–465 (2013).
Acknowledgements
The authors thank B. Hayes and B. Torok-Storb for gifting the hS5 cells, S. Adelmund for synthesizing and supplying BCN-OSu, E. Ruskowitz for useful discussion involving the DOX studies, as well as K. Anseth, D. Tirrell, S. Pun and B. Ratner for constructive comments during the preparation of this manuscript. The authors acknowledge support from S. Edgar at the University of Washington (UW) Mass Spectrometry Center, D. Prunkard at the UW Pathology Flow Cytometry Core Facility, and N. Peters and support from the NIH to the UW W. M. Keck Microscopy Center (S10 OD016240). This work was supported by a University of Washington Faculty Startup Grant (to C.A.D.) and a National Science Foundation CAREER Award (DMR 1652141, to C.A.D.).
Author information
Authors and Affiliations
Contributions
B.A.B. and C.A.D. conceived and designed the experiments. B.A.B., M.P.C., C.K.A. and J.A.S. performed the experiments. B.A.B. and C.A.D. analysed the data and prepared the figures. B.A.B. and C.A.D. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information
Supplementary information (PDF 5659 kb)
Rights and permissions
About this article
Cite this article
Badeau, B., Comerford, M., Arakawa, C. et al. Engineered modular biomaterial logic gates for environmentally triggered therapeutic delivery. Nature Chem 10, 251–258 (2018). https://doi.org/10.1038/nchem.2917
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchem.2917
This article is cited by
-
Recent progress in photoreactive crosslinkers in polymer network materials toward advanced photocontrollability
Polymer Journal (2024)
-
Irreversible light-activated SpyLigation mediates split-protein assembly in 4D
Nature Protocols (2024)
-
Dual-controlled guest release from coordination cages
Communications Chemistry (2024)
-
Smart and versatile biomaterials for cutaneous wound healing
Biomaterials Research (2023)
-
Tricolor visible wavelength-selective photodegradable hydrogel biomaterials
Nature Communications (2023)