Abstract
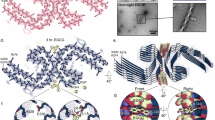
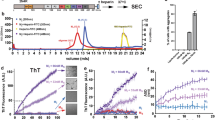
Aggregated tau protein is associated with over 20 neurological disorders, which include Alzheimer's disease. Previous work has shown that tau's sequence segments VQIINK and VQIVYK drive its aggregation, but inhibitors based on the structure of the VQIVYK segment only partially inhibit full-length tau aggregation and are ineffective at inhibiting seeding by full-length fibrils. Here we show that the VQIINK segment is the more powerful driver of tau aggregation. Two structures of this segment determined by the cryo-electron microscopy method micro-electron diffraction explain its dominant influence on tau aggregation. Of practical significance, the structures lead to the design of inhibitors that not only inhibit tau aggregation but also inhibit the ability of exogenous full-length tau fibrils to seed intracellular tau in HEK293 biosensor cells into amyloid. We also raise the possibility that the two VQIINK structures represent amyloid polymorphs of tau that may account for a subset of prion-like strains of tau.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Margittai, M. & Langen, R. Side chain-dependent stacking modulates tau filament structure. J. Biol. Chem. 281, 37820–37827 (2006).
von Bergen, M. et al. Assembly of τ protein into Alzheimer paired helical filaments depends on a local sequence motif (306VQIVYK311) forming β structure. Proc. Natl Acad. Sci. USA 97, 5129–5134 (2000).
Goedert, M., Eisenberg, D. S. & Crowther, R. A. Propagation of tau aggregates and neurodegeneration. Annu. Rev. Neurosci. 40, 189–210 (2017).
Schwarz, A. J. et al. Regional profiles of the candidate tau PET ligand recapitulate key features of Braak histopathological stages. Brain 139, 1539–1550 (2016).
Manczak, M. & Reddy, P. H. Abnormal interaction of oligomeric amyloid-β with phosphorylated tau: implications to synaptic dysfunction and neuronal damage. J. Alzheimers Dis. 36, 285–295 (2013).
Seward, M. E. et al. Amyloid-β signals through tau to drive ectopic neuronal cell cycle re-entry in Alzheimer's disease. J. Cell Sci. 126, 1278–1286 (2013).
Brier, M. R. et al. Tau and Aβ imaging, CSF measures, and cognition in Alzheimer's disease. Sci. Transl. Med. 8, 338ra366 (2016).
Kfoury, N., Holmes, B. B., Jiang, H., Holtzman, D. M. & Diamond, M. I. Trans-cellular propagation of tau aggregation by fibrillar species. J. Biol. Chem. 287, 19440–19451 (2012).
von Bergen, M. et al. Mutations of tau protein in frontotemporal dementia promote aggregation of paired helical filaments by enhancing local beta-structure. J. Biol. Chem., 276, 48165–48174 (2001).
Sawaya, M. R. et al. Atomic structures of amyloid cross-[bgr] spines reveal varied steric zippers. Nature 447, 453–457 (2007).
Sievers, S. A. et al. Structure-based design of non-natural amino-acid inhibitors of amyloid fibril formation. Nature 475, 96–100 (2011).
Zheng, J. et al. Macrocyclic β-sheet peptides that inhibit the aggregation of a tau-protein-derived hexapeptide. J. Am. Chem. Soc. 133, 3144–3157 (2011).
Shi, D., Nannenga, B. L., Iadanza, M. G. & Gonen, T. Three-dimensional electron crystallography of protein microcrystals. eLife 2, e01345 (2013).
Rodriguez, J. A. et al. Structure of the toxic core of [agr]-synuclein from invisible crystals. Nature 525, 486–490 (2015).
Rodriguez, J. A. & Gonen, T. in Methods in Enzymology Vol. 579 (ed. Crowther, R. A.) 369–392 (Academic, 2016).
Eisenberg, D. S. & Sawaya, M. R. Structural studies of amyloid proteins at the molecular level. Annu. Rev. Biochem. 86, 3.1–3.27 (2017).
Wood, S. J., Wetzel, R., Martin, J. D. & Hurle, M. R. Prolines and amyloidogenicity in fragments of the Alzheimer's peptide β/A4. Biochemistry 34, 724–730 (1995).
Moore, C. L. et al. Secondary nucleating sequences affect kinetics and thermodynamics of tau aggregation. Biochemistry 50, 10876–10886 (2011).
von Bergen, M. et al. Mutations of tau protein in frontotemporal dementia promote aggregation of paired helical filaments by enhancing local β-structure. J. Biol. Chem. 276, 48165–48174 (2001).
Margittai, M. & Langen, R. Fibrils with parallel in-register structure constitute a major class of amyloid fibrils: molecular insights from electron paramagnetic resonance spectroscopy. Q. Rev. Biophys. 41, 265–297 (2008).
Mirbaha, H., Holmes, B. B., Sanders, D. W., Bieschke, J. & Diamond, M. I. Tau trimers are the minimal propagation unit spontaneously internalized to seed intracellular aggregation. J. Biol. Chem., 290 14893–14903 (2015).
Sanders, D. W . et al. Distinct tau prion strains propagate in cells and mice and define different tauopathies. Neuron 82, 1271–1288 (2014).
Siddiqua, A. et al. Conformational basis for asymmetric seeding barrier in filaments of three- and four-repeat tau. J. Am. Chem. Soc. 134, 10271–10278 (2012).
Margittai, M. & Langen, R. Template-assisted filament growth by parallel stacking of tau. Proc. Natl Acad. Sci. USA 101, 10278–10283 (2004).
Fitzpatrick, A. W. P. et al. Cryo-EM structures of tau filaments from Alzheimer's disease. Nature 547, 185–190 (2017).
Kaufman, S. K. et al. Tau prion strains dictate patterns of cell pathology, progression rate, and regional vulnerability in vivo. Neuron 92, 796–812 (2016).
Wiltzius, J. J. W. et al. Molecular mechanisms for protein-encoded inheritance. Nat. Struct. Mol. Biol. 16, 973–978 (2009).
Kabsch, W. XDS. Acta Crystallogr. D 66, 125–132 (2010).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Murshudov, G. N., Vagin, A. A. & Dodson, E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53, 240–255 (1997).
Bricogne, G., B. E. et al. BUSTER 2.10.0 (Global Phasing Ltd, 2016).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr D 66, 486–501 (2010).
Acknowledgements
We thank M. Diamond for discussion and for gifting the monoclonal biosensor HEK293 cell-line that expressed tau 4R1N P301S-EYFP for our inhibitor assay, and H. Mirbaha for assistance and advice on conducting the biosensor seeding experiments and purifying K18 tau oligomers by gel filtration chromatography. We also thank awards 1R01 AG029430 and RF1 AG054022 from the National Institute on Aging, 1F32 NS095661 from the National Institute of Neurological Disorders and Stroke, A2016588F from the BrightFocus Foundation and HHMI and the Janelia Visiting Scientist Program for support.
Author information
Authors and Affiliations
Contributions
P.M.S. conceived and designed the experiments; P.M.S., D.R.B., J.A.R. and K.M. performed the experiments; P.M.S., D.R.B., M.R.S. and D.C. analysed data; and P.M.S., D.S.E. and T.G. co-wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information
Supplementary information (PDF 1963 kb)
Rights and permissions
About this article
Cite this article
Seidler, P., Boyer, D., Rodriguez, J. et al. Structure-based inhibitors of tau aggregation. Nature Chem 10, 170–176 (2018). https://doi.org/10.1038/nchem.2889
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchem.2889
This article is cited by
-
A novel transgenic mouse line with hippocampus-dominant and inducible expression of truncated human tau
Translational Neurodegeneration (2023)
-
Human tauopathy strains defined by phosphorylation in R1-R2 repeat domains of tau
Acta Neuropathologica Communications (2023)
-
Mechanisms and pathology of protein misfolding and aggregation
Nature Reviews Molecular Cell Biology (2023)
-
Tau Aggregation Inhibiting Peptides as Potential Therapeutics for Alzheimer Disease
Cellular and Molecular Neurobiology (2023)
-
Simulations of Amyloid-Forming Peptides in the Crystal State
The Protein Journal (2023)