Abstract
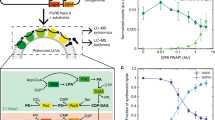
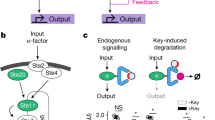
Genetic circuits and reaction cascades are of great importance for synthetic biology, biochemistry and bioengineering. An open question is how to maximize the modularity of their design to enable the integration of different reaction networks and to optimize their scalability and flexibility. One option is encapsulation within liposomes, which enables chemical reactions to proceed in well-isolated environments. Here we adapt liposome encapsulation to enable the modular, controlled compartmentalization of genetic circuits and cascades. We demonstrate that it is possible to engineer genetic circuit-containing synthetic minimal cells (synells) to contain multiple-part genetic cascades, and that these cascades can be controlled by external signals as well as inter-liposomal communication without crosstalk. We also show that liposomes that contain different cascades can be fused in a controlled way so that the products of incompatible reactions can be brought together. Synells thus enable a more modular creation of synthetic biology cascades, an essential step towards their ultimate programmability.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Carlson, E. D., Gan, R., Hodgman, C. E. & Jewett, M. C. Cell-free protein synthesis: applications come of age. Biotechnol. Adv. 30, 1185–1194 (2012).
Smith, M. T., Wilding, K. M., Hunt, J. M., Bennett, A. M. & Bundy, B. C. The emerging age of cell-free synthetic biology. FEBS Lett. 588, 2755–2761 (2014).
Hodgman, C. E. & Jewett, M. C. Cell-free synthetic biology: thinking outside the cell. Metab. Eng. 14, 261–269 (2012).
Miller, D. & Gulbis, J. Engineering protocells: prospects for self-assembly and nanoscale production-lines. Life 5, 1019–1053 (2015).
Shimizu, Y., Kuruma, Y., Ying, B.-W., Umekage, S. & Ueda, T. Cell-free translation systems for protein engineering. FEBS J. 273, 4133–4140 (2006).
Shin, J. & Noireaux, V. An E. coli cell-free expression toolbox: application to synthetic gene circuits and artificial cells. ACS Synth. Biol. 1, 29–41 (2012).
Takahashi, M. K. et al. Rapidly characterizing the fast dynamics of RNA genetic circuitry with cell-free transcription–translation (TX-TL) systems. ACS Synth. Biol. 4, 503–515 (2015).
Michener, J. K., Thodey, K., Liang, J. C. & Smolke, C. D. Applications of genetically-encoded biosensors for the construction and control of biosynthetic pathways. Metab. Eng. 14, 212–222 (2012).
Vamvakaki, V. & Chaniotakis, N. A. Pesticide detection with a liposome-based nano-biosensor. Biosens. Bioelectron. 22, 2848–2853 (2007).
Pardee, K. et al. Paper-based synthetic gene networks. Cell 159, 940–954 (2014).
Lentini, R. et al. Integrating artificial with natural cells to translate chemical messages that direct E. coli behaviour. Nat. Commun. 5, 4012 (2014).
Zemella, A., Thoring, L., Hoffmeister, C. & Kubick, S. Cell-free protein synthesis: pros and cons of prokaryotic and eukaryotic systems. Chem. Bio. Chem. 16, 2420–31 (2015).
Forster, A. C. & Church, G. M. Towards synthesis of a minimal cell. Mol. Syst. Biol. 2, 45 (2006).
Brea, R. J., Hardy, M. D. & Devaraj, N. K. Towards self-assembled hybrid artificial cells: novel bottom-up approaches to functional synthetic membranes. Chem. Pub. Soc. Euro. 21, 12564–12570 (2015).
Luisi, P. L., Ferri, F. & Stano, P. Approaches to semi-synthetic minimal cells: a review. Naturwissenschaften 93, 1–13 (2006).
Stano, P. & Luisi, P. L. Semi-synthetic minimal cells: origin and recent developments. Curr. Opin. Biotechnol. 24, 633–638 (2013).
Murtas, G., Kuruma, Y., Bianchini, P., Diaspro, A. & Luisi, P. L. Protein synthesis in liposomes with a minimal set of enzymes. Biochem. Biophys. Res. Commun. 363, 12–17 (2007).
Yu, W. et al. Synthesis of functional protein in liposome. J. Biosci. Bioeng. 92, 590–593 (2001).
Oberholzer, T., Nierhaus, K. H. & Luisi, P. L. Protein expression in liposomes. Biochem. Biophys. Res. Commun. 261, 238–241 (1999).
Noireaux, V. & Libchaber, A. A vesicle bioreactor as a step toward an artificial cell assembly. Proc. Natl Acad. Sci. USA 101, 17669–17674 (2004).
Stech, M. et al. Production of functional antibody fragments in a vesicle-based eukaryotic cell-free translation system. J. Biotechnol. 164, 220–231 (2012).
Weber, L. A, Feman, E. R. & Baglioni, C. A cell free system from HeLa cells active in initiation of protein synthesis. Biochemistry 14, 5315–5321 (1975).
Wimmer, E. Cell-free, de novo synthesis of poliovirus. Science. 254, 1647–1651, (1991).
Mikami, S., Masutani, M., Sonenberg, N., Yokoyama, S. & Imataka, H. An efficient mammalian cell-free translation system supplemented with translation factors. Protein Expr. Purif. 46, 348–357 (2006).
Mikami, S., Kobayashi, T., Masutani, M., Yokoyama, S. & Imataka, H. A human cell-derived in vitro coupled transcription/translation system optimized for production of recombinant proteins. Protein Expr. Purif. 62, 190–198 (2008).
Tan, C., Saurabh, S., Bruchez, M. P., Schwartz, R. & Leduc, P. Molecular crowding shapes gene expression in synthetic cellular nanosystems. Nat. Nanotechnol. 8, 602–608 (2013).
de Souza, T. P. et al. Encapsulation of ferritin, ribosomes, and ribo-peptidic complexes inside liposomes: insights into the origin of metabolism. Orig. Life Evol. Biosph. 42, 421–428 (2012).
de Souza, T. P., Fahr, A., Luisi, P. L. & Stano, P. Spontaneous encapsulation and concentration of biological macromolecules in liposomes: an intriguing phenomenon and its relevance in origins of life. J. Mol. Evol. 79, 179–192 (2014).
Caschera, F. & Noireaux, V. Integration of biological parts toward the synthesis of a minimal cell. Curr. Opin. Chem. Biol. 22, 85–91 (2014).
Stefureac, R., Long, Y. T., Kraatz, H. B., Howard, P. & Lee, J. S. Transport of α-helical peptides through α-hemolysin and aerolysin pores. Biochemistry 45, 9172–9179 (2006).
Gouaux, E., Hobaugh, M. & Song, L. α-Hemolysin, γ-hemolysin, and leukocidin from Staphylococcus aureus: distant in sequence but similar in structure. Protein Sci. 6, 2631–2635 (1997).
Selgrade, D. F., Lohmueller, J. J., Lienert, F. & Silver, P. A. Protein scaffold-activated protein trans-splicing in mammalian cells. 135, 7713–7719 (2013).
Tu, Y. et al. Mimicking the cell: bio-inspired functions of supramolecular assemblies. Chem. Rev. 116, 2023–2078 (2016).
Del Vecchio, D., Ninfa, A. J. & Sontag, E. D. Modular cell biology: retroactivity and insulation. Mol. Syst. Biol. 4, 161 (2008).
Caschera, F. et al. Programmed vesicle fusion triggers gene expression. Langmuir 27, 13082–13090 (2011).
Meyenberg, K., Lygina, A. S., van den Bogaart, G., Jahn, R. & Diederichsen, U. SNARE derived peptide mimic inducing membrane fusion. Chem. Commun. 47, 9405–9407 (2011).
Robson Marsden, H., Korobko, A. V., Zheng, T., Voskuhl, J. & Kros, A. Controlled liposome fusion mediated by SNARE protein mimics. Biomater. Sci. 1, 1046–1054 (2013).
Inglés-Prieto, Á. et al. Light-assisted small-molecule screening against protein kinases. Nat. Chem. Biol. 11, 952–954 (2015).
Boyden, E. S. A history of optogenetics: the development of tools for controlling brain circuits with light. F1000 Biol. Rep. 3, 11 (2011).
Hanczyc, M. M., Fujikawa, S. M. & Szostak, J. W. Experimental models of primitive cellular compartments: encapsulation, growth, and division. Science 302, 618–622 (2003).
Adamala, K. & Szostak, J. W. Competition between model protocells driven by an encapsulated catalyst. Nat. Chem. 5, 495–501 (2013).
Balaram, P. Synthesizing life. Curr. Sci. 85, 1509–1510 (2003).
Adamala, K. et al. Open questions in origin of life: experimental studies on the origin of nucleic acids and proteins with specific and functional sequences by a chemical synthetic biology approach. Comput. Struct. Biotechnol. J. 9, e201402004 (2014).
Ruiz-Mirazo, K., Briones, C. & de la Escosura, A. Prebiotic systems chemistry: new perspectives for the origins of life. Chem. Rev. 114, 285–366 (2014).
Glansdorff, N., Xu, Y. & Labedan, B. The last universal common ancestor: emergence, constitution and genetic legacy of an elusive forerunner. Biol. Direct 3, 29 (2008).
Woese, C. The universal ancestor. Proc. Natl Acad. Sci. USA 95, 6854–6859 (1998).
Theobald, D. L. A formal test of the theory of universal common ancestry. Nature 465, 219–222 (2010).
Spencer, A. C., Torre, P. & Mansy, S. S. The encapsulation of cell-free transcription and translation machinery in vesicles for the construction of cellular mimics. J. Vis. Exp. 80, e51304 (2013).
Adamala, K., Engelhart, A. E., Kamat, N. P., Jin, L. & Szostak, J. W. Construction of a liposome dialyzer for the preparation of high-value, small-volume liposome formulations. Nat. Protoc. 10, 927–938 (2015).
Shin, J. & Noireaux, V. Efficient cell-free expression with the endogenous E. coli RNA polymerase and sigma factor 70. J. Biol. Eng. 4, 8 (2010).
Lutz, R. & Bujard, H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 25, 1203–1210 (1997).
Guzman, L. M., Belin, D., Carson, M. J. & Beckwith, J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177, 4121–4130 (1995).
Sun, Z. Z. et al. Protocols for implementing an Escherichia coli based TX-TL cell-free expression system for synthetic biology. J. Vis. Exp. 79, e50762 (2013).
Acknowledgements
We thank E. Vasile and F. Chen for help with the SIM microscopy, and G. Paradis and K. Piatkevich for help with the flow-cytometry experiments. We thank N. Kamat and L. Jin for help with troubleshooting the DLS machine. We thank J. Szostak for sharing the liposome encapsulation formula. We thank V. Noireaux, A. Mershin and A. Engelhart for helpful discussions about cell-free TX/TL systems. E.S.B. acknowledges, for funding, the National Institutes of Health (NIH) 1U01MH106011, Jeremy and Joyce Wertheimer, NIH 1RM1HG008525, the Picower Institute Innovation Fund, NIH 1R01MH103910, NIH 1R01NS075421, National Science Foundation CBET 1053233, New York Stem Cell Foundation-Robertson Award and NIH Director's Pioneer Award 1DP1NS087724. D.A.M.-A. acknowledges support from the Janet and Sheldon Razin (1959) Fellowship.
Author information
Authors and Affiliations
Contributions
K.P.A. and D.A.M.-A. contributed equally to this work. K.P.A., D.A.M.-A. and K.R.G.-H. performed the experiments. K.P.A., D.A.M.-A. and E.S.B. designed experiments, analysed the data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
K.P.A., D.A.M.-A. and E.S.B. submitted a provisional patent application based on this work.
Supplementary information
Supplementary information
Supplementary information (PDF 1756 kb)
Rights and permissions
About this article
Cite this article
Adamala, K., Martin-Alarcon, D., Guthrie-Honea, K. et al. Engineering genetic circuit interactions within and between synthetic minimal cells. Nature Chem 9, 431–439 (2017). https://doi.org/10.1038/nchem.2644
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchem.2644
This article is cited by
-
DNA as a universal chemical substrate for computing and data storage
Nature Reviews Chemistry (2024)
-
Efficiency of transcription and translation of cell-free protein synthesis systems in cell-sized lipid vesicles with changing lipid composition determined by fluorescence measurements
Scientific Reports (2024)
-
Engineering cellular communication between light-activated synthetic cells and bacteria
Nature Chemical Biology (2023)
-
ANCA: artificial nucleic acid circuit with argonaute protein for one-step isothermal detection of antibiotic-resistant bacteria
Nature Communications (2023)
-
Synthetic cells with self-activating optogenetic proteins communicate with natural cells
Nature Communications (2022)