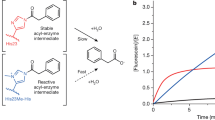
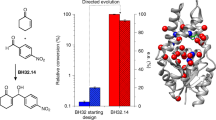
Abstract
The design of enzyme-like catalysts tests our understanding of sequence-to-structure/function relationships in proteins. Here we install hydrolytic activity predictably into a completely de novo and thermostable α-helical barrel, which comprises seven helices arranged around an accessible channel. We show that the lumen of the barrel accepts 21 mutations to functional polar residues. The resulting variant, which has cysteine–histidine–glutamic acid triads on each helix, hydrolyses p-nitrophenyl acetate with catalytic efficiencies that match the most-efficient redesigned hydrolases based on natural protein scaffolds. This is the first report of a functional catalytic triad engineered into a de novo protein framework. The flexibility of our system also allows the facile incorporation of unnatural side chains to improve activity and probe the catalytic mechanism. Such a predictable and robust construction of truly de novo biocatalysts holds promise for applications in chemical and biochemical synthesis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Schmid, A. et al. Industrial biocatalysis today and tomorrow. Nature 409, 258–268 (2001).
Nanda, V. & Koder, R. L. Designing artificial enzymes by intuition and computation. Nature Chem. 2, 15–24 (2010).
Hilvert, D. Design of protein catalysts. Annu. Rev. Biochem. 82, 447–470 (2013).
Woolfson, D. N. et al. De novo protein design: how do we expand into the universe of possible protein structures? Curr. Opin. Struct. Biol. 33, 16–26 (2015).
Kiss, G., Çelebi-Ölçüm, N., Moretti, R., Baker, D. & Houk, K. Computational enzyme design. Angew. Chem. Int. Ed. 52, 5700–5725 (2013).
Baker, D. An exciting but challenging road ahead for computational enzyme design. Protein Sci. 19, 1817–1819 (2010).
Richter, F., Leaver-Fay, A., Khare, S. D., Bjelic, S. & Baker, D. De novo enzyme design using Rosetta3. PLoS ONE 6, e19230 (2011).
Rajagopalan, S. et al. Design of activated serine-containing catalytic triads with atomic-level accuracy. Nature Chem. Biol. 10, 386–391 (2014).
Rothlisberger, D. et al. Kemp elimination catalysts by computational enzyme design. Nature 453, 190–195 (2008).
Jiang, L. et al. De novo computational design of retro-aldol enzymes. Science 319, 1387–1391 (2008).
Siegel, J. B. et al. Computational design of an enzyme catalyst for a stereoselective bimolecular Diels–Alder reaction. Science 329, 309–313 (2010).
Turner, N. J. Directed evolution drives the next generation of biocatalysts. Nature Chem. Biol. 5, 567–573 (2009).
Blomberg, R. et al. Precision is essential for efficient catalysis in an evolved Kemp eliminase. Nature 503, 418–421 (2013).
Kries, H., Blomberg, R. & Hilvert, D. De novo enzymes by computational design. Curr. Opin. Chem. Biol. 17, 221–228 (2013).
Giger, L. et al. Evolution of a designed retro-aldolase leads to complete active site remodeling. Nature Chem. Biol. 9, 494–498 (2013).
Preiswerk, N. et al. Impact of scaffold rigidity on the design and evolution of an artificial Diels–Alderase. Proc. Natl Acad. Sci. USA 111, 8013–8018 (2014).
Bolon, D. N. & Mayo, S. L. Enzyme-like proteins by computational design. Proc. Natl Acad. Sci. USA 98, 14274–14279 (2001).
Moroz, Y. S. et al. New tricks for old proteins: single mutations in a nonenzymatic protein give rise to various enzymatic activities. J. Am. Chem. Soc. 137, 14905–14911 (2015).
Richter, F. et al. Computational design of catalytic dyads and oxyanion holes for ester hydrolysis. J. Am. Chem. Soc. 134, 16197–16206 (2012).
Kaplan, J. & DeGrado, W. F. De novo design of catalytic proteins. Proc. Natl Acad. Sci. USA 101, 11566–11570 (2004).
Zastrow, M. L. & Pecoraro, V. L. Designing functional metalloproteins: from structural to catalytic metal sites. Coord. Chem. Rev. 257, 2565–2588 (2013).
Armstrong, C. T., Watkins, D. W. & Anderson, J. L. R. Constructing man-made enzymes for oxygen activation. J. Chem. Soc. Dalton Trans. 42, 3136–3150 (2013).
Broo, K. S., Brive, L., Ahlberg, P. & Baltzer, L. Catalysis of hydrolysis and transesterification reactions of p-nitrophenyl esters by a designed helix−loop−helix dimer. J. Am. Chem. Soc. 119, 11362–11372 (1997).
Nilsson, J. & Baltzer, L. Reactive-site design in folded-polypeptide catalysts—the leaving group pKa of reactive esters sets the stage for cooperativity in nucleophilic and general-acid catalysis. Chem. Eur. J. 6, 2214–2220 (2000).
Rufo, C. M. et al. Short peptides self-assemble to produce catalytic amyloids. Nature Chem. 6, 303–309 (2014).
Zastrow, M. L., Peacock, A. F. A., Stuckey, J. A. & Pecoraro, V. L. Hydrolytic catalysis and structural stabilization in a designed metalloprotein. Nature Chem. 4, 118–123 (2012).
Der, B. S., Edwards, D. R. & Kuhlman, B. Catalysis by a de novo zinc-mediated protein interface: implications for natural enzyme evolution and rational enzyme engineering. Biochemistry 51, 3933–3940 (2012).
Song, W. J. & Tezcan, F. A. A designed supramolecular protein assembly with in vivo enzymatic activity. Science 346, 1525–1528 (2014).
Wei, Y. & Hecht, M. H. Enzyme-like proteins from an unselected library of designed amino acid sequences. Prot. Eng. Des. Sel. 17, 67–75 (2004).
Woolfson, D. N. The design of coiled-coil structures and assemblies. Adv. Protein Chem. 70, 79–112 (2005).
Lupas, A. Coiled coils: new structures and new functions. Trends Biochem. Sci. 21, 375–382 (1996).
Calladine, C. R., Sharff, A. & Luisi, B. How to untwist an α-helix: structural principles of an α-helical barrel. J. Mol. Biol. 305, 603–618 (2001).
Woolfson, D. N., Bartlett, G. J., Bruning, M. & Thomson, A. R. New currency for old rope: from coiled-coil assemblies to α-helical barrels. Curr. Opin. Struct. Biol. 22, 432–441 (2012).
Zaccai, N. R. et al. A de novo peptide hexamer with a mutable channel. Nature Chem. Biol. 7, 935–941 (2011).
Thomson, A. R. et al. Computational design of water-soluble α-helical barrels. Science 346, 485–488 (2014).
Burton, A. J. et al. Accessibility, reactivity, and selectivity of side chains within a channel of de novo peptide assembly. J. Am. Chem. Soc. 135, 12524–12527 (2013).
Smith, A. J. T. et al. Structural reorganization and preorganization in enzyme active sites: comparisons of experimental and theoretically ideal active site geometries in the multistep serine esterase reaction cycle. J. Am. Chem. Soc. 130, 15361–15373 (2008).
Wood, C. W. et al. CCBuilder: an interactive web-based tool for building, designing and assessing coiled-coil protein assemblies. Bioinformatics 30, 3029–3035 (2014).
Fletcher, J. M. et al. A basis set of de novo coiled-coil peptide oligomers for rational protein design and synthetic biology. ACS Synth. Biol. 1, 240–250 (2012).
Huang, P.-S. et al. High thermodynamic stability of parametrically designed helical bundles. Science 346, 481–485 (2014).
Pellegrini-Calace, M., Maiwald, T. & Thornton, J. M. PoreWalker: a novel tool for the identification and characterization of channels in transmembrane proteins from their three-dimensional structure. PLoS Comput. Biol. 5, e1000440 (2009).
Bender, M. L., Kezdy, F. J. & Wedler, F. C. α-Chymotrypsin enzyme concentration and kinetics. J. Chem. Educ. 44, 84–87 (1967).
Kezdy, F. J. & Bender, M. L. The kinetics of the α-chymotrypsin-catalyzed hydrolysis of p-nitrophenyl acetate. Biochemistry 1, 1097–1106 (1962).
Faller, L. & Sturtevant, J. M. The kinetics of the α-chymotrypsin-catalyzed hydrolysis of p-nitrophenyl acetate in organic solvent–water mixtures. J. Biol. Chem. 241, 4825–4834 (1966).
Acknowledgements
A.J.B. thanks the Bristol Chemical Synthesis Centre for Doctoral Training funded by the Engineering and Physical Sciences Research Council (EP/G036764/1) and the University of Bristol for the provision of a PhD studentship. A.J.B., A.R.T., W.M.D. and D.N.W. are supported by the European Research Council (340764). D.N.W. holds a Royal Society Wolfson Research Merit Award. We thank the Diamond Light Source for access to beamlines I03, I04 and I24 (award MX-8922), and F. Thomas and members of the Woolfson group for helpful discussions.
Author information
Authors and Affiliations
Contributions
A.J.B., A.R.T. and D.N.W. designed the research. A.J.B. synthesized the peptides and the βMCys amino acid and performed the solution-phase biophysical analyses. A.J.B. and W.M.D. performed the kinetic experiments. A.J.B. and R.L.B. solved the X-ray crystal structures. A.J.B. and D.N.W. wrote the manuscript. All the authors analysed the data, and reviewed and contributed to the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information
Supplementary information (PDF 10149 kb)
Rights and permissions
About this article
Cite this article
Burton, A., Thomson, A., Dawson, W. et al. Installing hydrolytic activity into a completely de novo protein framework. Nature Chem 8, 837–844 (2016). https://doi.org/10.1038/nchem.2555
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchem.2555