Abstract
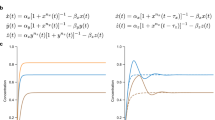
Analog molecular circuits can exploit the nonlinear nature of biochemical reaction networks to compute low-precision outputs with fewer resources than digital circuits. This analog computation is similar to that employed by gene-regulation networks. Although digital systems have a tractable link between structure and function, the nonlinear and continuous nature of analog circuits yields an intricate functional landscape, which makes their design counter-intuitive, their characterization laborious and their analysis delicate. Here, using droplet-based microfluidics, we map with high resolution and dimensionality the bifurcation diagrams of two synthetic, out-of-equilibrium and nonlinear programs: a bistable DNA switch and a predator–prey DNA oscillator. The diagrams delineate where function is optimal, dynamics bifurcates and models fail. Inverse problem solving on these large-scale data sets indicates interference from enzymatic coupling. Additionally, data mining exposes the presence of rare, stochastically bursting oscillators near deterministic bifurcations.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Rothemund, P. W. K. Folding DNA to create nanoscale shapes and patterns. Nature 440, 297–302 (2006).
Kuzyk, A. et al. DNA-based self-assembly of chiral plasmonic nanostructures with tailored optical response. Nature 483, 311–314 (2012).
Zhang, D. Y., Chen, S. X. & Yin, P. Optimizing the specificity of nucleic acid hybridization. Nature Chem. 4, 208–214 (2012).
Seelig, G. et al. Enzyme-free nucleic acid logic circuits. Science 314, 1585–1588 (2006).
Takinoue, M. et al. Experiments and simulation models of a basic computation element of an autonomous molecular computing system. Phys. Rev. E 78, 041921 (2008).
Katz, E. & Privman, V. Enzyme-based logic systems for information processing. Chem. Soc. Rev. 39, 1835–1857 (2010).
Genot, A. J., Bath, J. & Turberfield, A. J. Reversible logic circuits made of DNA. J. Am. Chem. Soc. 133, 20080–20083 (2011).
Qian, L. & Winfree, E. Scaling up digital circuit computation with DNA strand displacement cascades. Science 332, 1196–1201 (2011).
Orbach, R. et al. Logic reversibility and thermodynamic irreversibility demonstrated by DNAzyme-based Toffoli and Fredkin logic gates. Proc. Natl Acad. Sci. USA 109, 21228–21233 (2012).
Genot, A. J., Bath, J. & Turberfield, A. J. Combinatorial displacement of DNA strands: application to matrix multiplication and weighted sums. Angew. Chem. Int. Ed. 52, 1189–1192 (2013).
Daniel, R. et al. Synthetic analog computation in living cells. Nature 497, 619–623 (2013).
Sarpeshkar, R. Analog synthetic biology. Phil. Trans. R. Soc. A 372, 20130110 (2014).
Alon, U. An Introduction to Systems Biology (Chapman and Hall, 2007).
Hatakeyama, T. S. & Kaneko, K. Generic temperature compensation of biological clocks by autonomous regulation of catalyst concentration. Proc. Natl Acad. Sci. USA 109, 8109–8114 (2012).
Fujii, T. & Rondelez, Y. Predator–prey molecular ecosystems. ACS Nano 7, 27–34 (2013).
Montagne, K. et al. Programming an in vitro DNA oscillator using a molecular networking strategy. Mol. Syst. Biol. 7, 466 (2011).
Kim, J. & Winfree, E. Synthetic in vitro transcriptional oscillators. Mol. Syst. Biol. 7, 465 (2011).
Padirac, A., Fujii, T. & Rondelez, Y. Bottom-up construction of in vitro switchable memories. Proc. Natl Acad. Sci. USA 109, E3212–E3220 (2012).
Kim, J., White, K. S. & Winfree, E. Construction of an in vitro bistable circuit from synthetic transcriptional switches. Mol. Syst. Biol. 2, 68 (2006).
Qian, L., Winfree, E. & Bruck, J. Neural network computation with DNA strand displacement cascades. Nature 475, 368–372 (2011).
Chen, Y.-J. et al. Programmable chemical controllers made from DNA. Nature Nanotech. 8, 755–762 (2013).
Franco, E. et al. Timing molecular motion and production with a synthetic transcriptional clock. Proc. Natl Acad. Sci. USA 108, E784–E793 (2011).
Karzbrun, E. et al. Programmable on-chip DNA compartments as artificial cells. Science 345, 829–832 (2014).
Niederholtmeyer, H. et al. Rapid cell-free forward engineering of novel genetic ring oscillators. eLife e09771 (2015).
Epstein, I. R. & Pojman, J. A. An Introduction to Nonlinear Chemical Dynamics (Oxford Univ. Press, 1998).
Wei, B., Dai, M. & Yin, P. Complex shapes self-assembled from single-stranded DNA tiles. Nature 485, 623–626 (2012).
Niederholtmeyer, H., Stepanova, V. & Maerkl, S. J. Implementation of cell-free biological networks at steady state. Proc. Natl Acad. Sci. USA 110, 15985–15990 (2013).
Galas, J.-C., Haghiri-Gosnet, A.-M. & Estevez-Torres, A. A nanoliter-scale open chemical reactor. Lab Chip 13, 415–423 (2013).
Sugiura, H. et al. Pulse-density modulation control of chemical oscillation far from equilibrium in a droplet open-reactor system. Nature Commun. 7, 10212 (2016).
Hasatani, K. et al. High-throughput and long-term observation of compartmentalized biochemical oscillators. Chem. Commun. 49, 8090–8092 (2013).
Weitz, M. et al. Diversity in the dynamical behaviour of a compartmentalized programmable biochemical oscillator. Nature Chem. 6, 295–302 (2014).
Song, H., Chen, D. L. & Ismagilov, R. F. Reactions in droplets in microfluidic channels. Angew. Chem. Int. Ed. 45, 7336–7356 (2006).
Agresti, J. J. et al. Ultrahigh-throughput screening in drop-based microfluidics for directed evolution. Proc. Natl Acad. Sci. USA 107, 4004–4009 (2010).
Gardner, T. S., Cantor, C. R. & Collins, J. J. Construction of a genetic toggle switch in Escherichia coli. Nature 403, 339–342 (2000).
Baccouche, A. et al. Dynamic DNA-toolbox reaction circuits: a walkthrough. Methods 67, 234–249 (2014).
Crawford, J. D. Introduction to bifurcation theory. Rev. Mod. Phys. 63, 991–1037 (1991).
Dai, L. et al. Generic indicators for loss of resilience before a tipping point leading to population collapse. Science 336, 1175–1177 (2012).
Ma, R. et al. Small-number effects: a third stable state in a genetic bistable toggle switch. Phys. Rev. Lett. 109, 248107 (2012).
Huang, S. Reprogramming cell fates: reconciling rarity with robustness. Bioessays 31, 546–560 (2009).
Genot, A. J., Fujii, T. & Rondelez, Y. Computing with competition in biochemical networks. Phys. Rev. Lett. 109, 208102 (2012).
Hansen, N. & Ostermeier, A. Completely derandomized self-adaptation in evolution strategies. Evol. Comput. 9, 159–195 (2001).
Prindle, A. et al. Rapid and tunable post-translational coupling of genetic circuits. Nature 508, 387–391 (2014).
Van Roekel, H. W. H. et al. Automated design of programmable enzyme-driven DNA circuits. ACS Synth. Biol. 4, 735–745 (2015).
Decroly, O. & Goldbeter, A. Birhythmicity, chaos, and other patterns of temporal self-organization in a multiply regulated biochemical system. Proc. Natl Acad. Sci. USA 79, 6917–6921 (1982).
Tan, E. et al. Specific versus nonspecific isothermal DNA amplification through thermophilic polymerase and nicking enzyme activities. Biochemistry 47, 9987–9999 (2008).
Urabe, H. et al. Compartmentalization in a water-in-oil emulsion repressed the spontaneous amplification of RNA by Qβ replicase. Biochemistry 49, 1809–1813 (2010).
Tsimring, L. S. Noise in biology. Rep. Prog. Phys. 77, 026601 (2014).
Yamagata, A. et al. Overexpression, purification and characterization of RecJ protein from Thermus thermophilus HB8 and its core domain. Nucleic Acids Res. 29, 4617–4624 (2001).
Holtze, C. et al. Biocompatible surfactants for water-in-fluorocarbon emulsions. Lab Chip 8, 1632–1639 (2008).
Edelstein, A. D. et al. Advanced methods of microscope control using μManager software. J. Biol. Methods 1, e11 (2014).
Acknowledgements
This work was financially supported by the PHC Sakura program (project number 34171WG), implemented by the French Ministry of Foreign Affairs, the French Ministry Of Higher Education and Research and Japan Society for the Promotion of Science (JSPS), a Grant-in-Aid to Y.R. from the JSPS for Scientific Research on Innovative Areas ‘Synthetic Biology for Comprehension of Biomolecular Networks’ (project number 23119001), a l'Agence Nationale de la Recherche (ANR) Retour Postdoc grant (ANR-13-PDOC-0001) and a JSPS postdoctoral fellowship to A.J.G., and a PhD fellowship from Region Alsace to J.F.B. We thank H. Fujita and M. C. Tarhan for the loan of a microfluidic pump, A. Zadorin for discussions about the theory of bifurcations, K. Hasatani for preliminary work, E. Winfree for detailed comments on the manuscript and A. Estevez-Torres and Y. Tauran for expressing and purifying the exonuclease.
Author information
Authors and Affiliations
Contributions
A.J.G. designed the study, performed experiments, analysed the data and wrote the manuscript. A.B. performed experiments, improved the droplet chambers and contributed to experimental design and writing of the manuscript. R.S. developed the microfluidic platform. N.A.K. and N.B. designed, performed and analysed the fitting process. J.F.B. and V.T. synthesized the surfactant. V.T. and T.F. provided support with the droplet-based microfluidic platform. Y.R. conceived, designed and supervised the study, analysed data and wrote the manuscript. All the authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information
Supplementary information (PDF 10085 kb)
Supplementary information
Supplementary Movie 1 (AVI 3000 kb)
Supplementary information
Supplementary Movie 2 (MOV 4160 kb)
Supplementary information
Supplementary Movie 3 (MOV 3433 kb)
Supplementary information
Supplementary information (ZIP 29952 kb)
Rights and permissions
About this article
Cite this article
Genot, A., Baccouche, A., Sieskind, R. et al. High-resolution mapping of bifurcations in nonlinear biochemical circuits. Nature Chem 8, 760–767 (2016). https://doi.org/10.1038/nchem.2544
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchem.2544
This article is cited by
-
DNA-based programmable gate arrays for general-purpose DNA computing
Nature (2023)
-
Stochastic approximations of higher-molecular by bi-molecular reactions
Journal of Mathematical Biology (2023)
-
A deep artificial neural network powered by enzymes
Nature (2022)
-
Nonlinear decision-making with enzymatic neural networks
Nature (2022)
-
Programmable synthetic cell networks regulated by tuneable reaction rates
Nature Communications (2022)