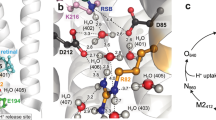
Abstract
Directional proton transport along ‘wires’ that feed biochemical reactions in proteins is poorly understood. Amino-acid residues with high pKa are seldom considered as active transport elements in such wires because of their large classical barrier for proton dissociation. Here, we use the light-triggered proton wire of the green fluorescent protein to study its ground-electronic-state proton-transport kinetics, revealing a large temperature-dependent kinetic isotope effect. We show that ‘deep’ proton tunnelling between hydrogen-bonded oxygen atoms with a typical donor–acceptor distance of 2.7–2.8 Å fully accounts for the rates at all temperatures, including the unexpectedly large value (2.5 × 109 s−1) found at room temperature. The rate-limiting step in green fluorescent protein is assigned to tunnelling of the ionization-resistant serine hydroxyl proton. This suggests how high-pKa residues within a proton wire can act as a ‘tunnel diode’ to kinetically trap protons and control the direction of proton flow.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Change history
09 June 2016
In the original version of this Article the caption for Fig. 5a contained incorrect information. was in fact shown by a black arrow. All versions of the Article have been corrected.
References
Kennis, J. T. M. et al. Uncovering the hidden ground state of green fluorescent protein. Proc. Natl Acad. Sci. USA 101, 17988–17993 (2004).
Kohen, A., Cannio, R., Bartolucci, S. & Klinman, J. P. Enzyme dynamics and hydrogen tunnelling in a thermophilic alcohol dehydrogenase. Nature 399, 496–499 (1999).
Masgrau, L. et al. Atomic description of an enzyme reaction dominated by proton tunneling. Science 312, 237–241 (2006).
Bhabha, G. et al. A dynamic knockout reveals that conformational fluctuations influence the chemical step of enzyme catalysis. Science 332, 234–238 (2011).
Knapp, M. J., Rickert, K. & Klinman, J. P. Temperature-dependent isotope effects in soybean lipoxygenase-1: correlating hydrogen tunneling with protein dynamics. J. Am. Chem. Soc. 124, 3865–3874 (2002).
Garczarek, F. & Gerwert, K. Functional waters in intraprotein proton transfer monitored by FTIR difference spectroscopy. Nature 439, 109–112 (2006).
Kuznetsov, A. M. & Ulstrup, J. Proton and hydrogen atom tunnelling in hydrolytic and redox enzyme catalysis. Can. J. Chem. 77, 1085–1096 (1999).
Hammes-Schiffer, S. & Stuchebrukhov, A. A. Theory of coupled electron and proton transfer reactions. Chem. Rev. 110, 6939–6960 (2010).
Antoniou, D. & Schwartz, S. D. Large kinetic isotope effects in enzymatic proton transfer and the role of substrate oscillations. Proc. Natl Acad. Sci. USA 94, 12360–12365 (1997).
Pu, J. Z., Gao, J. L. & Truhlar, D. G. Multidimensional tunneling, recrossing, and the transmission coefficient for enzymatic reactions. Chem. Rev. 106, 3140–3169 (2006).
Cui, Q. & Karplus, M. Promoting modes and demoting modes in enzyme-catalyzed proton transfer reactions: a study of models and realistic systems. J. Phys. Chem. B 106, 7927–7947 (2002).
Kiefer, P. M. & Hynes, J. T. Theoretical aspects of tunneling proton transfer reactions in a polar environment. J. Phys. Org. Chem. 23, 632–646 (2010).
Saraste, M. Oxidative phosphorylation at the fin de siecle. Science 283, 1488–1493 (1999).
Luecke, H., Richter, H. T. & Lanyi, J. K. Proton transfer pathways in bacteriorhodopsin at 2.3 angstrom resolution. Science 280, 1934–1937 (1998).
DeCoursey, T. E. Voltage-gated proton channels and other proton transfer pathways. Physiol. Rev. 83, 475–579 (2003).
Belevich, I., Verkhovsky, M. I. & Wikstrom, M. Proton-coupled electron transfer drives the proton pump of cytochrome c oxidase. Nature 440, 829–832 (2006).
Wraight, C. A. Chance and design—proton transfer in water, channels and bioenergetic proteins. Biochim. Biophys. Acta 1757, 886–912 (2006).
Nagle, J. F. & Morowitz, H. J. Molecular mechanisms for proton transport in membranes. Proc. Natl Acad. Sci. USA 75, 298–302 (1978).
Wikstrom, M., Sharma, V., Kaila, V. R. I., Hosler, J. P. & Hummer, G. New perspectives on proton pumping in cellular respiration. Chem. Rev. 115, 2196–2221 (2015).
Okamura, M. Y., Paddock, M. L., Graige, M. S. & Feher, G. Proton and electron transfer in bacterial reaction centers. Biochim. Biophys. Acta 1458, 148–163 (2000).
Nagano, S. & Poulos, T. L. Crystallographic study on the dioxygen complex of wild-type and mutant cytochrome P450cam—implications for the dioxygen activation mechanism. J. Biol. Chem. 280, 31659–31663 (2005).
Truhlar, D. G. Tunneling in enzymatic and nonenzymatic hydrogen transfer reactions. J. Phys. Org. Chem. 23, 660–676 (2010).
Van Thor, J. J., Georgiev, G. Y., Towrie, M. & Sage, J. T. Ultrafast and low barrier motions in the photoreactions of the green fluorescent protein. J. Biol. Chem. 280, 33652–33659 (2005).
Braun-Sand, S., Strajbl, M. & Warshel, A. Studies of proton translocations in biological systems: simulating proton transport in carbonic anhydrase by EVB-based models. Biophys. J. 87, 2221–2239 (2004).
Zhang, H., Sun, Q., Li, Z., Nanbu, S. & Smith, S. S. First principle study of proton transfer in the green fluorescent protein (GFP): ab initio PES in a cluster model. Comp. Theor. Chem. 990, 185–193 (2012).
Pomes, R. & Roux, B. Structure and dynamics of a proton wire: a theoretical study of H+ translocation along the single-file water chain in the gramicidin A channel. Biophys. J. 71, 19–39 (1996).
Vendrell, O., Gelabert, R., Moreno, M. & Lluch, J. M. A potential energy function for heterogeneous proton-wires. Ground and photoactive states of the proton-wire in the green fluorescent protein. J. Chem. Theory Comput. 4, 1138–1150 (2008).
Benabbas, A., Salna, B., Sage, J. T. & Champion, P. M. Deep proton tunneling in the electronically adiabatic and non-adiabatic limits: comparison of the quantum and classical treatment of donor–acceptor motion in a protein environment. J. Chem. Phys. 142, 114101 (2015).
Garcia-Viloca, M., Gao, J., Karplus, M. & Truhlar, D. G. How enzymes work: analysis by modern rate theory and computer simulations. Science 303, 186–195 (2004).
Kamerlin, S. C. L. & Warshel, A. At the dawn of the 21st century: is dynamics the missing link for understanding enzyme catalysis? Proteins 78, 1339–1375 (2010).
Chattoraj, M., King, B. A., Bublitz, G. U. & Boxer, S. G. Ultra-fast excited state dynamics in green fluorescent protein: multiple states and proton transfer. Proc. Natl Acad. Sci. USA 93, 8362–8367 (1996).
Nesheim, J. C. & Lipscomb, J. D. Large kinetic isotope effects in methane oxidation catalyzed by methane monooxygenase: evidence for C–H bond cleavage in a reaction cycle intermediate. Biochemistry 35, 10240–10247 (1996).
Roston, D., Islam, Z. & Kohen, A. Isotope effects as probes for enzyme catalyzed hydrogen-transfer reactions. Molecules 18, 5543–5567 (2013).
Sutcliffe, M. J. et al. Hydrogen tunnelling in enzyme-catalysed H-transfer reactions: flavoprotein and quinoprotein systems. Phil. Trans. R. Soc. Lond. B 361, 1375–1386 (2006).
Hay, S. & Scrutton, N. S. Good vibrations in enzyme-catalysed reactions. Nature Chem. 4, 161–168 (2012).
Hay, S., Sutcliffe, M. J. & Scrutton, N. S. Promoting motions in enzyme catalysis probed by pressure studies of kinetic isotope effects. Proc. Natl Acad. Sci. USA 104, 507–512 (2007).
Hatcher, E., Soudackov, A. V. & Hammes-Schiffer, S. Proton-coupled electron transfer in soybean lipoxygenase. J. Am. Chem. Soc. 126, 5763–5775 (2004).
Thompson, L. M. et al. Analytical harmonic vibrational frequencies for the green fluorescent protein computed with ONIOM: chromophore mode character and its response to environment. J. Chem. Theory Comput. 10, 751–766 (2014).
Andrews, B. T., Gosavi, S., Finke, J. M., Onuchic, J. N. & Jennings, P. A. The dual-basin landscape in GFP folding. Proc. Natl Acad. Sci. USA 105, 12283–12288 (2008).
Novak, A. Hydrogen bonding in solids. Correlation of spectroscopic and crystallographic data. Struct. Bond. 18, 177–216 (1974).
Premont-Schwarz, M., Barak, T., Pines, D., Nibbering, E. T. J. & Pines, E. Ultrafast excited-state proton-transfer reaction of 1-naphthol-3,6-disulfonate and several 5-substituted 1-naphthol derivatives. J. Phys. Chem. B 117, 4594–4603 (2013).
Shinobu, A., Gottfried, J. P., Schierbeek, A. J. & Agmon, N. Visualizing proton antenna in a high-resolution green fluorescent protein structure. J. Am. Chem. Soc. 132, 11093–11102 (2010).
van Thor, J. J., Zanetti, G., Ronayne, K. L. & Towrie, M. Structural events in the photocycle of green fluorescent protein. J. Phys. Chem. B 109, 16099–16108 (2005).
Walrafen, G. E. & Pugh, E. Raman combinations and stretching overtones from water, heavy water, and NaCl in water at shifts to ca. 7000 cm–1. J. Solution Chem. 33, 81–97 (2004).
Laptenok, S. P. et al. Complete proton transfer cycle in GFP and its T203V and S205V mutants. Angew. Chem. Int. Ed. 54, 9303–9307 (2015).
Shu, X. et al. An alternative excited-state proton transfer pathway in green fluorescent protein variant S205V. Protein Sci. 16, 2703–2710 (2007).
Okamura, M. Y. & Feher, G. Proton transfer in reaction centers from photosynthetic bacteria. Annu. Rev. Biochem. 61, 861–896 (1992).
Pisliakov, A. V., Sharma, P. K., Chu, Z. T., Haranczyk, M. & Warshel, A. Electrostatic basis for the unidirectionality of the primary proton transfer in cytochrome c oxidase. Proc. Natl Acad. Sci. USA 105, 7726–7731 (2008).
Goyal, P., Yang, S. & Cui, Q. Microscopic basis for kinetic gating in cytochrome c oxidase: insights from QM/MM analysis. Chem. Sci. 6, 826–841 (2015).
Xu, J. & Voth, G. A. Computer simulation of explicit proton translocation in cytochrome c oxidase: the D-pathway. Proc. Natl Acad. Sci. USA 102, 6795–6800 (2005).
DeCoursey, T. E. & Hosler, J. Philosophy of voltage-gated proton channels. J. R. Soc. Interface 11, 20130799 (2014).
Sagnella, D. E. & Straub, J. E. Directed energy “funneling” mechanism for heme cooling following ligand photolysis or direct excitation in solvated carbonmonoxy myoglobin. J. Phys. Chem. B 105, 7057–7063 (2001).
Baradaran, R., Berrisford, J. M., Minhas, G. S. & Sazanov, L. A. Crystal structure of the entire respiratory complex I. Nature 494, 443–448 (2013).
Ginovska-Pangovska, B. et al. Molecular dynamics study of the proposed proton transport pathways in FeFe -hydrogenase. Biochim. Biophys. Acta. 1837, 131–138 (2014).
Saito, K., Rutherford, A. W. & Ishikita, H. Mechanism of proton-coupled quinone reduction in photosystem II. Proc. Natl Acad. Sci. USA 110, 954–959 (2014).
Van Thor, J. J., Pierik, A. J., Nugteren-Roodzant, I., Xie, A. H. & Hellingwerf, K. J. Characterization of the photoconversion of green fluorescent protein with FTIR spectroscopy. Biochemistry 37, 16915–16921 (1998).
Yu, A. C., Ye, X., Ionascu, D., Cao, W. X. & Champion, P. M. Two-color pump–probe laser spectroscopy instrument with picosecond time-resolved electronic delay and extended scan range. Rev. Sci. Instrum. 76, 114301 (2005).
Acknowledgements
This work was supported by National Science Foundation awards CHE-1243948 (P.M.C) and CHE-1026369 (P.M.C. and J.T.S.) and EPSRC award EP/I003304/1 (J.v.T.). The authors thank M. Bearpark and L. Thompson for providing the ONIOM files associated with the GFP normal mode calculation and A. Fitzpatrick for preparing the E222D mutant. A. McClelland and B. Kellner are also thanked for early contributions to this work.
Author information
Authors and Affiliations
Contributions
A.B. and B.S. carried out the experiments, performed the data analysis, and helped to develop the theoretical models. In doing so, they contributed equally to this work. P.M.C., J.v.T. and J.T.S. conceived the experiments and developed the theoretical approaches. P.M.C. wrote the paper with input from the other authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information
Supplementary information (PDF 2358 kb)
Rights and permissions
About this article
Cite this article
Salna, B., Benabbas, A., Sage, J. et al. Wide-dynamic-range kinetic investigations of deep proton tunnelling in proteins. Nature Chem 8, 874–880 (2016). https://doi.org/10.1038/nchem.2527
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchem.2527
This article is cited by
-
Positive functional synergy of structurally integrated artificial protein dimers assembled by Click chemistry
Communications Chemistry (2019)
-
Water assisted biomimetic synergistic process and its application in water-jet rewritable paper
Nature Communications (2018)