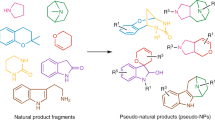
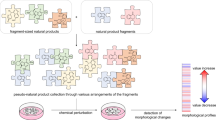
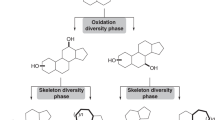
Abstract
Natural products and their molecular frameworks have a long tradition as valuable starting points for medicinal chemistry and drug discovery. Recently, there has been a revitalization of interest in the inclusion of these chemotypes in compound collections for screening and achieving selective target modulation. Here we discuss natural-product-inspired drug discovery with a focus on recent advances in the design of synthetically tractable small molecules that mimic nature's chemistry. We highlight the potential of innovative computational tools in processing structurally complex natural products to predict their macromolecular targets and attempt to forecast the role that natural-product-derived fragments and fragment-like natural products will play in next-generation drug discovery.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Lee, M. L. & Schneider, G. Scaffold architecture and pharmacophoric properties of natural products and trade drugs: application in the design of natural product-based combinatorial libraries. J. Comb. Chem. 3, 284–289 (2001).
Du, L. et al. Crowdsourcing natural products discovery to access uncharted dimensions of fungal metabolite diversity. Angew. Chem. Int. Ed. 53, 804–809 (2014).
Bauer, A. & Brönstrup, M. Industrial natural product chemistry for drug discovery and development. Nat. Prod. Rep. 31, 35–60 (2014).
Brown, D. G., Lister, T. & May-Dracka, T. L. New natural products as new leads for antibacterial drug discovery. Bioorg. Med. Chem. Lett. 24, 413–418 (2014).
Scheepstra, M. et al. A natural-product switch for a dynamic protein interface. Angew. Chem. Int. Ed. 53, 6443–6448 (2014).
Hattum, H. v. & Waldmann, H. Biology-oriented synthesis: harnessing the power of evolution. J. Am. Chem. Soc. 136, 11853–11859 (2014).
Harvey, A. L., Edrada-Ebel, R. & Quinn, R. J. The re-emergence of natural products for drug discovery in the genomics era. Nature Rev. Drug Discov. 14, 111–129 (2015).
Koch, M. A. et al. Charting biologically relevant chemical space: a structural classification of natural products (SCONP). Proc. Natl Acad. Sci. USA 102, 17272–17277 (2005).
Eschenbrenner-Lux, V., Ku¨chler, P., Ziegler, S., Kumar, K. & Waldmann, H. An enantioselective inverse-electron-demand imino Diels-Alder reaction. Angew. Chem. Int. Ed. 53, 2134–2137 (2014).
Zimmermann, T. J. et al. Biology-oriented synthesis of a tetrahydroisoquinoline-based compound collection targeting microtubule polymerization. ChemBioChem 14, 295–300 (2013).
Narayan, R., Bauer, J. O., Strohmann, C., Antonchick, A. P. & Waldmann, H. Catalytic enantioselective synthesis of functionalized tropanes reveals novel inhibitors of hedgehog signaling. Angew. Chem. Int. Ed. 52, 12892–12896 (2013).
Lowe, D. B. Drug discovery: combichem all over again. Nature Chem. 6, 851–852 (2014).
Molinari, G. Natural products in drug discovery: present status and perspectives. Adv. Exp. Med. Biol. 655, 13–27 (2009).
Koehn, F. E. & Carter, G. T. The evolving role of natural products in drug discovery. Nature Rev. Drug Discov. 4, 206–220 (2005).
Harvey, A. L. Natural products in drug discovery. Drug Discov. Today 13, 894–901 (2008).
Patridge, E., Gareiss, P., Kinch, M. S. & Hoyer, D. An analysis of FDA-approved drugs: natural products and their derivatives. Drug Discov. Today 21, 204–207 (2015).
Basu, S. & Waldmann, H. Polymer supported synthesis of a natural product-inspired oxepane library. Bioorg. Med. Chem. 22, 4430–3333 (2014).
Huigens, R. W. III et al. A ring-distortion strategy to construct stereochemically complex and structurally diverse compounds from natural products. Nature Chem. 5, 195–202 (2013).
Keller, N. P. Translating biosynthetic gene clusters into fungal armor and weaponry. Nature Chem. Biol. 11, 671–677 (2015).
Medema, M. H. & Fischbach, M. A. Computational approaches to natural product discovery. Nature Chem. Biol. 11, 639–648 (2015).
Kim, E., Moore, B. S. & Yoon, Y. J. Reinvigorating natural product combinatorial biosynthesis with synthetic biology. Nature Chem. Biol. 11, 649–659 (2015).
Morrison, K. C. & Hergenrother, P. J. Natural products as starting points for the synthesis of complex and diverse compounds. Nat. Prod. Rep. 31, 6–14 (2014).
Henkel, T., Brunne, R. M., Müller, H. & Reichel, F. Statistical investigation into the structural complementarity of natural products and synthetic compounds. Angew. Chem. Int. Ed. 38, 643–647 (1999).
Ertl, P., Jelfs, S., Muhlbacher, J., Schuffenhauer, A. & Selzer, P. Quest for the rings. In silico exploration of ring universe to identify novel bioactive heteroaromatic scaffolds. J. Med. Chem. 49, 4568–4573 (2006).
Akbulut, Y. et al. (−)-Englerin A is a potent and selective activator of TRPC4 and TRPC5 calcium channels. Angew. Chem. Int. Ed. 54, 3787–3791 (2015).
Schuster, D., Laggner, C. & Langer, T. Why drugs fail — a study on side effects in new chemical entities. Curr. Pharm. Des. 11, 3545–3559 (2005).
Hopkins, A. L., Keseru, G. M., Leeson, P. D., Rees, D. C. & Reynolds, C. H. The role of ligand efficiency metrics in drug discovery. Nature Rev. Drug Discov. 13, 105–121 (2014).
Murray, C. W. & Rees, D. C. The rise of fragment-based drug discovery. Nature Chem. 1, 187–192 (2009).
Grabowski, K., Baringhaus, K. H. & Schneider, G. Scaffold diversity of natural products: inspiration for combinatorial library design. Nat. Prod. Rep. 25, 892–904 (2008).
Barelier, S. et al. Increasing chemical space coverage by combining empirical and computational fragment screens. ACS Chem. Biol. 9, 1528–1535 (2014).
Reker, D. et al. Revealing the macromolecular targets of complex natural products. Nature Chem. 6, 1072–1078 (2014).
Reutlinger, M., Rodrigues, T., Schneider, P. & Schneider, G. Multi-objective molecular de novo design by adaptive fragment prioritization. Angew. Chem. Int. Ed. 53, 4244–4248 (2014).
Parkinson, E. I. et al. Deoxynybomycins inhibit mutant DNA gyrase and rescue mice infected with fluoroquinolone-resistant bacteria. Nature Commun. 6, 6947 (2015).
Lipinski, C. A. Drug-like properties and the causes of poor solubility and poor permeability. J. Pharmacol. Toxicol. Meth. 44, 235–249 (2000).
Congreve, M., Carr, R., Murray, C. & Jhoti, H. A rule of three for fragment-based lead discovery? Drug Discov. Today 8, 876–877 (2003).
Baell, J., Congreve, M., Leeson, P. & Abad-Zapatero, C. Ask the experts: past, present and future of the rule of five. Future Med. Chem. 5, 745–752 (2013).
Ntie-Kang, F., Lifongo, L. L., Judson, P. N., Sippl, W. & Efange, S. M. How “drug-like” are naturally occurring anti-cancer compounds? J. Mol. Model. 20, 2069 (2014).
Kenny, P. W. & Montanari, C. A. Inflation of correlation in the pursuit of drug-likeness. J. Comput. Aided Mol. Des. 27, 1–13 (2013).
Johnstone, C. Medicinal chemistry matters — a call for discipline in our discipline. Drug Discov. Today 17, 538–543 (2012).
Quinn, R. J. et al. Developing a druglike natural product library. J. Nat. Prod. 71, 464–468 (2008).
Antonchick, A. P. et al. Highly enantioselective synthesis and cellular evaluation of spirooxindoles inspired by natural products. Nature Chem. 2, 735–740 (2010).
Svenda, J. et al. Biology-oriented synthesis of a withanolide-inspired compound collection reveals novel modulators of hedgehog signalling. Angew. Chem. Int. Ed. 54, 5596–5602 (2015).
Schmid, F., Jessen, H. J., Burch, P. & Gademann, K. Truncated militarinone fragments identified by total chemical synthesis induce neurite outgrowth. Med. Chem. Comm. 4, 135–139 (2013).
DeLorbe, J. E., Clements, J. H., Whiddon, B. B. & Martin, S. F. Thermodynamic and structural effects of macrocyclic constraints in protein−ligand interactions. ACS Med. Chem. Lett. 1, 448–452 (2010).
Giordanetto, F. & Kihlberg, J. Macrocyclic drugs and clinical candidates: What can medicinal chemists learn from their properties? J. Med. Chem. 57, 278–295 (2014).
Zhang, J. et al. Diversity-oriented synthesis of Lycopodium alkaloids inspired by the hidden functional group pairing pattern. Nature Commun. 5, 4614 (2014).
Beckmann, H. S. et al. A strategy for the diversity-oriented synthesis of macrocyclic scaffolds using multidimensional coupling. Nature Chem. 5, 861–867 (2013).
Karageorgis, G., Warriner, S. & Nelson, A. Efficient discovery of bioactive scaffolds by activity-directed synthesis. Nature Chem. 6, 872–876 (2014).
McLeod, M. C. et al. Probing chemical space with alkaloid-inspired libraries. Nature Chem. 6, 133–140 (2014).
Elumalai, N., Berg, A., Natarajan, K., Scharow, A. & Berg, T. Nanomolar inhibitors of the transcription factor STAT5b with high selectivity over STAT5a. Angew. Chem. Int. Ed. 54, 4756–4763 (2015).
Larsson, J., Gottfries, J., Muresan, S. & Backlund, A. ChemGPS-NP: tuned for navigation in biologically relevant chemical space. J. Nat. Prod. 70, 789–794 (2007).
Miyao, T., Reker, D., Schneider, P., Funatsu, K. & Schneider, G. Chemography of natural product space. Planta Med. 81, 429–435 (2015).
Reutlinger, M. & Schneider, G. Nonlinear dimensionality reduction and mapping of compound libraries for drug discovery. J. Mol. Graph. Model. 34, 108–117 (2012).
Bon, R. S. & Waldmann, H. Bioactivity-guided navigation of chemical space. Acc. Chem. Res. 43, 1103–1114 (2010).
Lachance, H., Wetzel, S., Kumar, K. & Waldman, H. Charting, navigating, and populating natural product chemical space for drug discovery. J. Med. Chem. 55, 5989–6001 (2012).
Takayama, H. et al. Discovery of inhibitors of the Wnt and Hedgehog signaling pathways through the catalytic enantioselective synthesis of an iridoid-inspired compound collection. Angew. Chem. Int. Ed. 52, 12404–12408 (2013).
Dakas, P. Y. et al. Discovery of neuritogenic compound classes inspired by natural products. Angew. Chem. Int. Ed. 52, 9576–9581 (2013).
Voigt, T. et al. A natural product inspired tetrahydropyran collection yields mitosis modulators that synergistically target CSE1L and tubulin. Angew. Chem. Int. Ed. 52, 410–414 (2013).
Bennani, Y. L. Drug discovery in the next decade: innovation needed ASAP. Drug Discov. Today 16, 779–792 (2011).
Wetzel, S. et al. Interactive exploration of chemical space with Scaffold Hunter. Nature Chem. Biol. 5, 581–583 (2009).
Renner, S. et al. Bioactivity-guided mapping and navigation of chemical space. Nature Chem. Biol. 5, 585–592 (2009).
Over, B. et al. Natural-product-derived fragments for fragment-based ligand discovery. Nature Chem. 5, 21–28 (2013).
Bickerton, G. R., Paolini, G. V., Besnard, J., Muresan, S. & Hopkins, A. L. Quantifying the chemical beauty of drugs. Nature Chem. 4, 90–98 (2012).
Nickel, J. et al. SuperPred: update on drug classification and target prediction. Nucleic Acids Res. 42, W26–W31 (2014).
Keiser, M. J. et al. Predicting new molecular targets for known drugs. Nature 462, 175–181 (2009).
Keiser, M. J. et al. Relating protein pharmacology by ligand chemistry. Nature Biotechnol. 25, 197–206 (2007).
Gfeller, D. et al. SwissTargetPrediction: a web server for target prediction of bioactive small molecules. Nucleic Acids Res. 42, W32–W38 (2014).
Reutlinger, M., Rodrigues, T., Schneider, P. & Schneider, G. Combining on-chip synthesis of a focused combinatorial library with computational target prediction reveals imidazopyridine GPCR ligands. Angew. Chem. Int. Ed. 53, 582–585 (2014).
Antolin, A. A., Jalencas, X., Yelamos, J. & Mestres, J. Identification of pim kinases as novel targets for PJ34 with confounding effects in PARP biology. ACS Chem. Biol. 7, 1962–1967 (2012).
Hopkins, A. L. Network pharmacology: the next paradigm in drug discovery. Nature Chem. Biol. 4, 682–690 (2008).
Lounkine, E. et al. Large-scale prediction and testing of drug activity on side-effect targets. Nature 486, 361–367 (2012).
Rollinger, J. M. Accessing target information by virtual parallel screening — the impact on natural product research. Phytochem. Lett. 2, 53–58 (2009).
Do, Q. T. et al. Reverse pharmacognosy: identifying biological properties for plants by means of their molecule constituents: application to meranzin. Planta Med. 73, 1235–1240 (2007).
Vuorinen, A., Nashev, L. G., Odermatt, A., Rollinger, J. M. & Schuster, D. Pharmacophore model refinement for 11β-hydroxysteroid dehydrogenase inhibitors: search for modulators of intracellular glucocorticoid concentrations. Mol. Inf. 33, 15–25 (2014).
Bauer, J. et al. Discovery of depsides and depsidones from lichen as potent inhibitors of microsomal prostaglandin E2 synthase-1 using pharmacophore models. ChemMedChem 7, 2077–2081 (2012).
Rollinger, J. M. et al. In silico target fishing for rationalized ligand discovery exemplified on constituents of Ruta graveolens. Planta Med. 75, 195–204 (2009).
Schuster, D. et al. Morphinans and isoquinolines: acetylcholinesterase inhibition, pharmacophore modeling, and interaction with opioid receptors. Bioorg. Med. Chem. 18, 5071–5080 (2010).
Fakhrudin, N. et al. Computer-aided discovery, validation, and mechanistic characterization of novel neolignan activators of peroxisome proliferator-activated receptor gamma. Mol. Pharmacol. 77, 559–566 (2010).
Atanasov, A. G. et al. Honokiol: a non-adipogenic PPARγ agonist from nature. Biochim. Biophys. Acta 1830, 4813–4819 (2013).
Arrowsmith, C. H. et al. The promise and peril of chemical probes. Nature Chem. Biol. 11, 536–541 (2015).
Lagunin, A., Stepanchikova, A., Filimonov, D. & Poroikov, V. PASS: prediction of activity spectra for biologically active substances. Bioinformatics 16, 747–748 (2000).
Lagunin, A., Filimonov, D. & Poroikov, V. Multi-targeted natural products evaluation based on biological activity prediction with PASS. Curr. Pharm. Des. 16, 1703–1717 (2010).
Filimonov, D., Poroikov, V., Borodina, Y. & Gloriozova, T. Chemical similarity assessment through multilevel neighbourhoods of atoms: definition and comparison with the other descriptors. J. Chem. Inf. Comput. Sci. 39, 666–670 (1999).
Chen, X. et al. Drug–target interaction prediction: databases, web servers and computational models. Brief. Bioinform. http://dx.doi.org/10.1093/bib/bbv066 (2015).
Liu, X. et al. Predicting targeted polypharmacology for drug repositioning and multi-target drug discovery. Curr. Med. Chem. 20, 1646–1661 (2013).
Pahikkala, T. et al. Toward more realistic drug–target interaction predictions. Brief. Bioinform. 16, 325–337 (2015).
Mousavian, Z. & Masoudi-Nejad, A. Drug–target interaction prediction via chemogenomic space: learning-based methods. Expert Opin. Drug Metab. Toxicol. 10, 1273–1287 (2014).
Yamanishi, Y. Chemogenomic approaches to infer drug-target interaction networks. Meth. Mol. Biol. 939, 97–113 (2013).
Rupp, M. et al. Machine learning estimates of natural product conformational energies. PLoS Comput. Biol. 10, e1003400 (2014).
Reker, D., Rodrigues, T., Schneider, P. & Schneider, G. Identifying the macromolecular targets of de novo-designed chemical entities through self-organizing map consensus. Proc. Natl Acad. Sci. USA 111, 4067–4072 (2014).
Schneider, G., Reker, D., Rodrigues, T. & Schneider, P. Coping with polypharmacology by computational medicinal chemistry. Chimia 68, 648–653 (2014).
Reutlinger, M. et al. Chemically advanced template search (CATS) for scaffold-hopping and prospective target prediction for 'orphan' molecules. Mol. Inf. 32, 133–138 (2013).
Reker, D. et al. Deorphaning pyrrolopyrazines as potent multi-target antimalarial agents. Angew. Chem. Int. Ed. 53, 7079–7084 (2014).
Rodrigues, T., Reker, D., Kunze, J., Schneider, P. & Schneider, G. Revealing the macromolecular targets of fragment-like natural products. Angew. Chem. Int. Ed. 54, 10516–10520 (2015).
Imming, P. Molecular targets of natural drug substances: idiosyncrasies and preferences. Planta Med. 76, 1794–1801 (2010).
Koeberle, A. & Werz, O. Multi-target approach for natural products in inflammation. Drug Discov. Today 19, 1871–1882 (2014).
Geldenhuys, W. J. & Van der Schyf, C. J. Rationally designed multi-targeted agents against neurodegenerative diseases. Curr. Med. Chem. 20, 1662–1672 (2013).
Schmeller, T., Sauerwein, M., Sporer, F., Wink, M. & Muller, W. E. Binding of quinolizidine alkaloids to nicotinic and muscarinic acetylcholine receptors. J. Nat. Prod. 57, 1316–1319 (1994).
Kirchmair, J. et al. Predicting drug metabolism: experiment and/or computation? Nature Rev. Drug Discov. 14, 387–404 (2015).
Gertsch, J. Botanical drugs, synergy, and network pharmacology: forth and back to intelligent mixtures. Planta Med. 77, 1086–1098 (2011).
Mak, L. et al. Anti-cancer drug development: computational strategies to identify and target proteins involved in cancer metabolism. Curr. Pharm. Des. 19, 532–577 (2013).
de Sá Alves, F. R., Barreiro, E. J. & Fraga, C. A. From nature to drug discovery: the indole scaffold as a 'privileged structure'. Mini Rev. Med. Chem. 9, 782–793 (2009).
Venkatraman, P. Specificity in the interaction of natural products with their target proteins — a biochemical and structural insight. Mini Rev. Med. Chem. 10, 540–549 (2010).
Mishra, B. B. & Tiwari, V. K. Natural products: an evolving role in future drug discovery. Eur. J. Med. Chem. 46, 4769–4807 (2011).
Jayaseelan, K. V., Moreno, P., Truszkowski, A., Ertl, P. & Steinbeck, C. Natural product-likeness score revisited: an open-source, open-data implementation. BMC Bioinform. 13, 106 (2012).
Roche, O. et al. Development of a virtual screening method for identification of “frequent hitters” in compound libraries. J. Med. Chem. 45, 137–142 (2002).
Coan, K. E. & Shoichet, B. K. Stoichiometry and physical chemistry of promiscuous aggregate-based inhibitors. J. Am. Chem. Soc. 130, 9606–9612 (2008).
Seidler, J., McGovern, S. L., Doman, T. N. & Shoichet, B. K. Identification and prediction of promiscuous aggregating inhibitors among known drugs. J. Med. Chem. 46, 4477–4486 (2003).
Baell, J. & Walters, M. A. Chemistry: chemical con artists foil drug discovery. Nature 513, 481–483 (2014).
Petrova, T., Chuprina, A., Parkesh, R. & Pushechnikov, A. Structural enrichment of HTS compounds from available commercial libraries. Med. Chem. Comm. 3, 571–579 (2012).
Walters, W. P., Stahl, M. T. & Murcko, M. A. Virtual screening — an overview. Drug Discov. Today 3, 160–178 (1998).
Duan, D., Doak, A. K., Nedyalkova, L. & Shoichet, B. K. Colloidal aggregation and the in vitro activity of traditional Chinese medicines. ACS Chem. Biol. 10, 978–988 (2015).
Wishart, D. S. et al. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 36, D901–D906 (2008).
Bento, A. P. et al. The ChEMBL bioactivity database: an update. Nucleic Acids Res. 42, D1083–D1090 (2014).
Chen, C. Y. TCM Database@Taiwan: the world's largest traditional Chinese medicine database for drug screening in silico. PLoS ONE 6, e15939 (2011).
Rishton, G. M. Reactive compounds and in vitro false positives in HTS. Drug Discov. Today 2, 382–384 (1997).
Hann, M. et al. Strategic pooling of compounds for high-throughput screening. J. Chem. Inf. Comput. Sci. 39, 897–902 (1999).
Baell, J. B. & Holloway, G. A. New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J. Med. Chem. 53, 2719–2740 (2010).
Lagorce, D., Sperandio, O., Baell, J. B., Miteva, M. A. & Villoutreix, B. O. FAF-Drugs3: a web server for compound property calculation and chemical library design. Nucleic Acids Res. 43, W200–W207 (2015).
Jost, C., Nitsche, C., Scholz, T., Roux, L. & Klein, C. D. Promiscuity and selectivity in covalent enzyme inhibition: a systematic study of electrophilic fragments. J. Med. Chem. 57, 7590–7599 (2014).
Schneider, P., Röthlisberger, M., Reker, D. & Schneider, G. Spotting and designing promiscuous ligands for drug discovery. Chem. Commun. 52, 1135–1138 (2016).
Clemons, P. A. et al. Small molecules of different origins have distinct distributions of structural complexity that correlate with protein-binding profiles. Proc. Natl Acad. Sci. USA 107, 18787–18792 (2010).
Kell, D. B. Finding novel pharmaceuticals in the systems biology era using multiple effective drug targets, phenotypic screening and knowledge of transporters: where drug discovery went wrong and how to fix it. FEBS J. 280, 5957–5980 (2013).
Rollinger, J. M., Stuppner, H. & Langer, T. Virtual screening for the discovery of bioactive natural products. Prog. Drug Res. 65, 213–249 (2008).
Harvey, A. L., Edrada-Ebel, R. & Quinn, R. J. The re-emergence of natural products for drug discovery in the genomics era. Nature Rev. Drug Discov. 14, 111–129 (2015).
Cragg, G. M., Grothaus, P. G. & Newman, D. J. New horizons for old drugs and drug leads. J. Nat. Prod. 77, 703–723 (2014).
Schuster, D. & Wolber, G. Identification of bioactive natural products by pharmacophore-based virtual screening. Curr. Pharm. Des. 16, 1666–1681 (2010).
Jacoby, E. & Mozzarelli, A. Chemogenomic strategies to expand the bioactive chemical space. Curr. Med. Chem. 16, 4374–4381 (2009).
Hübel, K., Lessmannm, T. & Waldmann, H. Chemical biology — identification of small molecule modulators of cellular activity by natural product inspired synthesis. Chem. Soc. Rev. 37, 1361–1374 (2008).
Kumar, K. & Waldmann, H. Synthesis of natural product inspired compound collections. Angew. Chem. Int. Ed. 48, 3224–3242 (2009).
Hirai, G. Mimicking/extracting structure and functions of natural products: synthetic approaches that address unexplored needs in chemical biology. Chem. Rec. 15, 445–456 (2015).
Thomas, G. L. & Johannes, C. W. Natural product-like synthetic libraries. Curr. Opin. Chem. Biol. 15, 516–522 (2011).
Gupta, P. K., Barone, G., Gurley, B. J., Fifer, E. K. & Hendrickson, H. P. Hydrastine pharmacokinetics and metabolism after a single oral dose of goldenseal (Hydrastis canadensis) to humans. Drug Metab. Dispos. 43, 534–552 (2015).
Bier, D., Thiel, P., Briels, J. & Ottmann, C. Stabilization of protein–protein interactions in chemical biology and drug discovery. Prog. Biophys. Mol. Biol. 119, 10–19 (2015).
Klenner, A., Hartenfeller, M., Schneider, P. & Schneider, G. 'Fuzziness' in pharmacophore-based virtual screening and de novo design. Drug Discov. Today Technol. 7, e237–e244 (2010).
Lewell, X. Q., Judd, D. B., Watson, S. P. & Hann, M. M. RECAP — retrosynthetic combinatorial analysis procedure: a powerful new technique for identifying privileged molecular fragments with useful applications in combinatorial chemistry. J. Chem. Inf. Comput. Sci. 38, 511–522 (1998).
Kusari, S. et al. Tramadol — a true natural product? Angew. Chem. Int. Ed. Engl. 53, 12073–12076 (2014).
Talalay, P., De Long, M. J. & Prochaska, H. J. Identification of a common chemical signal regulating the induction of enzymes that protect against chemical carcinogenesis. Proc. Natl Acad. Sci. USA 85, 8261–8265 (1988).
Aptula, A. O. & Roberts, D. W. Mechanistic applicability domains for nonanimal-based prediction of toxicological end points: general principles and application to reactive toxicity. Chem. Res. Toxicol. 19, 1097–1105 (2006).
Amslinger, S. The tunable functionality of α,β-unsaturated carbonyl compounds enables their differential application in biological systems. ChemMedChem 5, 351–356 (2010).
Avonto, C. et al. An NMR spectroscopic method to identify and classify thiol-trapping agents: revival of Michael acceptors for drug discovery? Angew. Chem. Int. Ed. 50, 467–471 (2011).
Acknowledgements
We thank K.-H. Altmann for invaluable advice and discussion. This study was financially supported by the OPO Foundation, Zürich.
Author information
Authors and Affiliations
Contributions
All authors contributed equally.
Corresponding author
Ethics declarations
Competing interests
P.S. and G.S. are the founders of inSili.com LLC, Zürich. All the other authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Rodrigues, T., Reker, D., Schneider, P. et al. Counting on natural products for drug design. Nature Chem 8, 531–541 (2016). https://doi.org/10.1038/nchem.2479
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchem.2479
This article is cited by
-
Synthesis of 2,9-dihydropyrano[2,3-b]-indoles via intramolecular oxa-6π-electrocyclization reaction from synthesized 3-allylideneindolin-2-one intermediates
Chemical Papers (2024)
-
Inhibitory effects of quercetin and resveratrol and their sulfonamide derivatives on the carbonic anhydrase activity: spectroscopic studies of the binding process
Chemical Papers (2024)
-
Network pharmacology‑based investigation of potential targets of triptonodiol acting on non-small-cell lung cancer
European Journal of Medical Research (2023)
-
Variational autoencoder-based chemical latent space for large molecular structures with 3D complexity
Communications Chemistry (2023)
-
Modeling the expansion of virtual screening libraries
Nature Chemical Biology (2023)