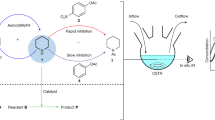
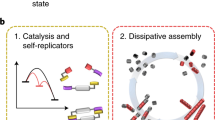
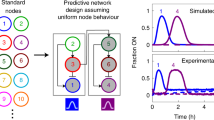
Abstract
Life is sustained by complex systems operating far from equilibrium and consisting of a multitude of enzymatic reaction networks. The operating principles of biology's regulatory networks are known, but the in vitro assembly of out-of-equilibrium enzymatic reaction networks has proved challenging, limiting the development of synthetic systems showing autonomous behaviour. Here, we present a strategy for the rational design of programmable functional reaction networks that exhibit dynamic behaviour. We demonstrate that a network built around autoactivation and delayed negative feedback of the enzyme trypsin is capable of producing sustained oscillating concentrations of active trypsin for over 65 h. Other functions, such as amplification, analog-to-digital conversion and periodic control over equilibrium systems, are obtained by linking multiple network modules in microfluidic flow reactors. The methodology developed here provides a general framework to construct dissipative, tunable and robust (bio)chemical reaction networks.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Bray, D. Protein molecules as computational elements in living cells. Nature 376, 307–312 (1995).
Koshland, D. E., Goldbeter, A. & Stock, J. B. Amplification and adaptation in regulatory and sensory systems. Science 217, 220–225 (1982).
Kholodenko, B. N. Cell-signalling dynamics in time and space. Nature Rev. Mol. Cell Biol. 7, 165–176 (2006).
Ferrell, J. E., Tsai, T. Y. & Yang, C. Q. O. Modeling the cell cycle: why do certain circuits oscillate? Cell 144, 874–885 (2011).
Yashin, V. V. & Balazs, A. C. Pattern formation and shape changes in self-oscillating polymer gels. Science 314, 798–801 (2006).
Stuart, M. A. C. et al. Emerging applications of stimuli-responsive polymer materials. Nature Mater. 9, 101–113 (2011).
Debnath, S., Roy, S. & Ulijn, R. V. Peptide nanofibers with dynamic instability through nonequilibrium biocatalytic assembly. J. Am. Chem. Soc. 135, 16789–16792 (2013).
Elowitz, M. B. & Leibler, S. A synthetic oscillatory network of transcriptional regulators. Nature 403, 335–338 (2000).
Hasty, J., McMillen, D. & Collins, J. J. Engineered gene circuits. Nature 420, 224–230 (2002).
Kim, J. & Winfree, E. Synthetic in vitro transcriptional oscillators. Mol. Syst. Biol. 7, 465 (2011).
Franco, E. et al. Timing molecular motion and production with a synthetic transcriptional clock. Proc. Natl Acad. Sci. USA 108, 784–793 (2011).
Chirieleison, S. M., Allen, P. B., Simpson, Z. B., Ellington, A. D. & Chen, X. Pattern transformation with DNA circuits. Nature Chem. 5, 1000–1005 (2013).
Fujii, T. & Rondelez, Y. Predator–prey molecular ecosystems. ACS Nano 7, 27–34 (2013).
Montagne, K., Plasson, R., Sakai, Y., Fujii, T. & Rondelez, Y. Programming an in vitro DNA oscillator using a molecular networking strategy. Mol. Syst. Biol. 7, 466 (2011).
Epstein, I. R. & Showalter, K. Nonlinear chemical dynamics: oscillations, patterns, and chaos. J. Phys. Chem. 100, 13132–13147 (1996).
Horváth, J., Szalai, I. & De Kepper, P. An experimental design method leading to chemical Turing patterns. Science 324, 772–775 (2009).
De Kepper, P., Epstein, I. R. & Kustin, K. A. Systematically designed homogeneous oscillating reaction: the arsenite-iodate-chlorite system. J. Am. Chem. Soc. 103, 2133–2134 (1981).
Taylor, A. F., Tinsley, M. R., Wang, F., Huang, Z. & Showalter, K. Dynamical quorum sensing and synchronization in large populations of chemical oscillators. Science 323, 614–617 (2009).
Yoshida, R., Takahashi, T., Yamaguchi, T. & Ichijo, H. Self-oscillating gel. J. Am. Chem. Soc. 118, 5134–5135 (1996).
Boekhoven, J. et al. Catalytic control over supramolecular gel formation. Nature Chem. 5, 433–437 (2013).
Boekhoven, J. et al. Dissipative self-assembly of a molecular gelator by using a chemical fuel. Angew. Chem. Int. Ed. 49, 4825–4828 (2010).
Carnall, J. M. A. et al. Mechanosensitive self-replication driven by self-organization. Science 327, 1502–1506 (2010).
He, X. et al. Synthetic homeostatic materials with chemo-mechano-chemical self-regulation. Nature 487, 214–218 (2012).
Gerdts, C. J., Sharoyan, D. E. & Ismagilov, R. F. A synthetic reaction network: chemical amplification using nonequilibrium autocatalytic reactions coupled in time. J. Am. Chem. Soc. 126, 6327–6331 (2004).
Warren, S. C., Guney-Altay, O. & Grzybowski, B. A. Responsive and nonequilibrium nanomaterials. J. Phys. Chem. Lett. 3, 2103–2111 (2012).
Goodwin, B. C. Oscillatory behavior in enzymatic control processes. Adv. Enzyme Regul. 3, 425–438 (1965).
Novak, B. & Tyson, J. J. Design principles of biochemical oscillators. Nature Rev. Mol. Cell Biol. 9, 981–991 (2008).
Kurin-Csörgei, K., Epstein, I. R. & Orbán, M. Systematic design of chemical oscillators using complexation and precipitation equilibria. Nature 433, 139–142 (2005).
Boissonade, J. & De Kepper, P. Transitions from bistability to limit cycle oscillations. Theoretical analysis and experimental evidence in an open chemical system. J. Phys. Chem. 84, 501–506 (1980).
Aubel, D. & Fussenegger, M. Watch the clock—engineering biological systems to be on time. Curr. Opin. Genet. Dev. 20, 634–643 (2010).
Abita, J-P., Delaage, M. & Lazdunsk, M. Mechanism of activation of trypsinogen—role of 4 N-terminal aspartyl residues. Eur. J. Biochem. 8, 314–324 (1969).
Seely, J. H. & Benoiton, N. L. Effect of N-methylation and chain length on kinetic constants of trypsin substrates epsilon-N-methyllysine and homolysine derivatives as substrates. Can. J. Biochem. Cell B 48, 1122–1131 (1970).
Barkai, N. & Leibler, S. Robustness in simple biochemical networks. Nature 387, 913–917 (1997).
Lucasius, C. B. & Kateman, G. Understanding and using genetic algorithms—Part 1. Concepts, properties and context. Chemometr. Intell. Lab. Sys. 19, 1–33 (1993).
Priftis, D. & Tirrell, M. Phase behavior and complex coacervation of aqueous polypeptide solutions. Soft Matter 8, 9396–9405 (2012).
Acknowledgements
The authors thank H. Adams for help with peptide synthesis. This work was supported by the European Research Council (ERC, advanced grant 246812 Intercom, to W.T.S.H.), the Netherlands Organization for Scientific Research (NWO, VICI grant 700.10.44, to W.T.S.H.), a Marie Curie Intra-European Fellowship (grant 300519, to S.N.S.) and funding from the Dutch Ministry of Education, Culture and Science (Gravity programme 024.001.035).
Author information
Authors and Affiliations
Contributions
W.T.S.H. supervised the research. S.N.S., A.S.Y.W. and W.T.S.H. planned the project and designed experiments. S.N.S. designed the oscillating network, and synthesized and optimized all compounds. S.N.S., A.S.Y.W., S.G.J.P. and J.G. performed experiments and analysed data. S.N.S., A.S.Y.W. and R.M.M. built and refined the model and wrote the required scripts for analysis. A.S.Y.W., R.M.M., H.W.H.R. and T.F.A.G. performed computational simulations. S.N.S., A.S.Y.W., S.G.J.P., T.F.A.G., and W.T.S.H. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information
Supplementary information (PDF 3720 kb)
Rights and permissions
About this article
Cite this article
Semenov, S., Wong, A., van der Made, R. et al. Rational design of functional and tunable oscillating enzymatic networks. Nature Chem 7, 160–165 (2015). https://doi.org/10.1038/nchem.2142
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchem.2142
This article is cited by
-
Interlinking spatial dimensions and kinetic processes in dissipative materials to create synthetic systems with lifelike functionality
Nature Nanotechnology (2024)
-
A catalytically active oscillator made from small organic molecules
Nature (2023)
-
Photoswitchable gating of non-equilibrium enzymatic feedback in chemically communicating polymersome nanoreactors
Nature Chemistry (2023)
-
Feedback-controlled hydrogels with homeostatic oscillations and dissipative signal transduction
Nature Nanotechnology (2022)
-
Standardized excitable elements for scalable engineering of far-from-equilibrium chemical networks
Nature Chemistry (2022)