Abstract
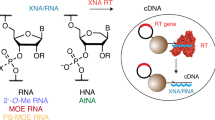
A plausible process for non-enzymatic RNA replication would greatly simplify models of the transition from prebiotic chemistry to simple biology. However, all known conditions for the chemical copying of an RNA template result in the synthesis of a complementary strand that contains a mixture of 2′–5′ and 3′–5′ linkages, rather than the selective synthesis of only 3′–5′ linkages as found in contemporary RNA. Here we show that such backbone heterogeneity is compatible with RNA folding into defined three-dimensional structures that retain molecular recognition and catalytic properties and, therefore, would not prevent the evolution of functional RNAs such as ribozymes. Moreover, the same backbone heterogeneity lowers the melting temperature of RNA duplexes that would otherwise be too stable for thermal strand separation. By allowing copied strands to dissociate, this heterogeneity may have been one of the essential features that allowed RNA to emerge as the first biopolymer.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Gilbert, W. The RNA world. Nature 319, 618 (1986).
Joyce, G. F. RNA evolution and the origins of life. Nature 338, 217–224 (1989).
Joyce, G. F. & Orgel, L. E. in The RNA World 2nd edn (eds R. F. Gesteland & J. F. Atkins) 49–77 (Cold Spring Harbor Laboratory Press, 1999).
Ban, N., Nissen, P., Hansen, J., Moore, P. B. & Steitz, T. A. The complete atomic structure of the large ribosomal subunit at 2.4 Ångstrom resolution. Science 289, 905–920 (2000).
Nissen, P., Hansen, J., Ban, N., Moore, P. B. & Steitz, T. A. The structural basis of ribosome activity in peptide bond synthesis. Science 289, 920–930 (2000).
Powner, M. W., Gerland, B. & Sutherland, J. D. Synthesis of activated pyrimidine ribonucleotides in prebiotically plausible conditions. Nature 459, 239–242 (2009).
Powner, M. W., Sutherland, J. D. & Szostak, J. W. Chemoselective multicomponent one-pot assembly of purine precursors in water. J. Am. Chem. Soc. 132, 16677–16688 (2010).
Powner, M. W. & Sutherland, J. D. Phosphate-mediated interconversion of ribo- and arabino-configured prebiotic nucleotide intermediates. Angew. Chem. Int. Ed. 49, 4641–4643 (2010).
Ferris, J. P. Jr, Hill, A. R., Liu, R. & Orgel, L. E. Synthesis of long prebiotic oligomers on mineral surfaces. Nature 381, 59–61 (1996).
Kanavarioti, A., Monnard, P-A. & Deamer, D. W. Eutectic phases in ice facilitate nonenzymatic nucleic acid synthesis. Astrobiology 1, 271–281 (2001).
Hill, A. J., Orgel, L. & Wu, T. The limits of template-directed synthesis with nucleoside-5′-phosphoro(2-methyl)imidazolides. Origins Life Evol. Biospheres 23, 285–290 (1993).
Orgel, L. E. Prebiotic chemistry and the origin of the RNA world. Crit. Rev. Biochem. Mol. Biol. 39, 99–123 (2004).
Deck, C., Jauker, M. & Richert, C. Efficient enzyme-free copying of all four nucleobases templated by immobilized RNA. Nature Chem. 3, 603–608 (2011).
Szostak, J. W. The eightfold path to non-enzymatic RNA replication. J. Syst. Chem. 3, 2 doi:10.1186/1759-2208-3-2 (2012).
Joyce, G. F., Schwartz, A. W., Miller, S. L. & Orgel, L. E. The case for an ancestral genetic system involving simple analogues of the nucleotides. Proc. Natl Acad. Sci. USA 84, 4398–4402 (1987).
Egholm, M. et al. PNA hybridizes to complementary oligonucleotides obeying the Watson–Crick hydrogen-bonding rules. Nature 365, 566–568 (1993).
Richert, C., Roughton, A. L. & Benner, S. A. Nonionic analogs of RNA with dimethylene sulfone bridges. J. Am. Chem. Soc. 118, 4518–4531 (1996).
Eschenmoser, A. Chemical etiology of nucleic acid structure. Science 284, 2118–2124 (1999).
Chaput, J. C. & Switzer, C. Nonenzymatic oligomerization on templates containing phosphodiester-linked acyclic glycerol nucleic acid analogues. J. Mol. Evol. 51, 464–470 (2000).
Bean, H. D., Anet, F. A. L., Gould, I. R. & Hud, N. V. Glyoxylate as a backbone linkage for a prebiotic ancestor of RNA. Origins Life Evol. Biospheres 36, 39–63 (2006).
Chen, J. J., Cai, X. & Szostak, J. W. N2′→P3′ phosphoramidate glycerol nucleic acid as a potential alternative genetic system. J. Am. Chem. Soc. 131, 2119–2121 (2009).
Engelhart, A. E. & Hud, N. V. Primitive genetic polymers. Cold Spring Harbor Perspect. Biol. 2, doi:10.1101/cshperspect.a002196 (2010).
Usher, D. A. & McHale, A. H. Hydrolytic stability of helical RNA – selective advantage for the natural 3′,5′-bond. Proc. Natl Acad. Sci. USA 73, 1149–1153 (1976).
Bridson, P. K. & Orgel, L. E. Catalysis of accurate poly(C)-directed synthesis of 3′–5′-linked oligoguanylates by Zn2+. J. Mol. Biol. 144, 567–577 (1980).
Inoue, T. & Orgel, L. E. Oligomerization of (guanosine-5′-phosphor)-2-methylimidazole on poly(C) – an RNA-polymerase model. J. Mol. Biol. 162, 201–217 (1982).
Rohatgi, R., Bartel, D. P. & Szostak, J. W. Nonenzymatic, template-directed ligation of oligoribonucleotides is highly regioselective for the formation of 3′–5′ phosphodiester bonds. J. Am. Chem. Soc. 118, 3340–3344 (1996).
Ekland, E. H. & Bartel, D. P. RNA-catalysed RNA polymerization using nucleoside triphosphates. Nature 382, 373–376 (1996).
Johnston, W. K., Unrau, P. J., Lawrence, M. S., Glasner, M. E. & Bartel, D. P. RNA-catalyzed RNA polymerization: accurate and general RNA-templated primer extension. Science 292, 1319–1325 (2001).
Prakash, T. P., Roberts, C. & Switzer, C. Activity of 2′,5′-linked RNA in the template-directed oligomerization of mononucleotides. Angew. Chem. Int. Ed. Engl. 36, 1522–1523 (1997).
Trevino, S. G., Zhang, N., Elenko, M. P., Luptak, A. & Szostak, J. W. Evolution of functional nucleic acids in the presence of nonheritable backbone heterogeneity. Proc. Natl Acad. Sci. USA 108, 13492–13497 (2011).
Giannaris, P. A. & Damha, M. J. Oligoribonucleotides containing 2′,5′-phosphodiester linkages exhibit binding selectivity for 3′,5′-RNA over 3′,5′-ssDNA. Nucleic Acids Res. 21, 4742–4749 (1993).
Wasner, M. et al. Physicochemical and biochemical properties of 2′,5′-linked RNA and 2′,5′-RNA:3′,5′-RNA ‘hybrid’ duplexes. Biochemistry 37, 7478–7486 (1998).
Burgstaller, P. & Famulok, M. Isolation of RNA aptamers for biological cofactors by in-vitro selection. Angew. Chem. Int. Ed. Engl. 33, 1084–1087 (1994).
Fan, P., Suri, A. K., Fiala, R., Live, D. & Patel, D. J. Molecular recognition in the FMN–RNA aptamer complex. J. Mol. Biol. 258, 480–500 (1996).
Birikh, K. R., Heaton, P. A. & Eckstein, F. The structure, function and application of the hammerhead ribozyme. Eur. J. Biochem. 245, 1–16 (1997).
Hiraki, S. & Orgel, L. E. Oligonucleotide synthesis catalyzed by the Zn2+ ion. J. Am. Chem. Soc. 97, 3532–3533 (1975).
Burlina, F., Fourrey, J., Lefort, V. & Favre, A. Cleavage activity of a hammerhead ribozyme domain containing 2′,5′-phosphodiester linkages. Tetrahedron Lett. 40, 4559–4562 (1999).
Shih, I. & Been, M. D. Ribozyme cleavage of a 2′,5′-phosphodiester linkage: mechanism and a restricted divalent metal-ion requirement. RNA 5, 1140–1148 (1999).
Lorsch, J. R., Bartel, D. P. & Szostak, J. W. Reverse transcriptase reads through a 2′–5′ linkage and a 2′-thiophosphate in a template. Nucleic Acids Res. 23, 2811–2814 (1995).
Pinheiro, V. B. et al. Synthetic genetic polymers capable of heredity and evolution. Science 336, 341–344 (2012).
Ricardo, A. & Szostak, J. W. Life on earth. Sci Am 301, 54–61 (2009).
Budin, I. & Szostak, J. W. Expanding roles for diverse physical phenomena during the origin of life. Annu. Rev. Biophys. 39, 245–263 (2010).
Budin, I. & Szostak, J. W. Physical effects underlying the transition from primitive to modern cell membranes. Proc. Natl Acad. Sci. USA 108, 5249–5254 (2011).
Qu, X. G. & Chaires, J. B. Analysis of drug–DNA binding data. Method. Enzymol. 321, 353–369 (2000).
Acknowledgements
J.W.S. is an Investigator of the Howard Hughes Medical Institute (HHMI). A.E.E. is supported by an appointment to the NASA Postdoctoral Program, administered by Oak Ridge Associated Universities through a contract with NASA. M.W.P. was an HHMI Research Associate. This work was supported in part through NSF Grant CHE-0809413 to J.W.S. We thank J. Craig Blain for oligonucleotide mass spectrometry, K. Adamala for advice with RNA melting experiments and N. Prywes for helpful discussions and assistance with figure preparation.
Author information
Authors and Affiliations
Contributions
All authors contributed to the design of the experiments and to writing the paper. Experiments were conducted by A.E.E. and M.W.P.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information
Supplementary information (PDF 706 kb)
Rights and permissions
About this article
Cite this article
Engelhart, A., Powner, M. & Szostak, J. Functional RNAs exhibit tolerance for non-heritable 2′–5′ versus 3′–5′ backbone heterogeneity. Nature Chem 5, 390–394 (2013). https://doi.org/10.1038/nchem.1623
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchem.1623
This article is cited by
-
Nonenzymatic Template-Directed Primer Extension Using 2′-3′ Cyclic Nucleotides Under Wet-Dry Cycles
Origins of Life and Evolution of Biospheres (2023)
-
Phosphorylation in liquid sulfur dioxide under prebiotically plausible conditions
Communications Chemistry (2022)
-
The role of sugar-backbone heterogeneity and chimeras in the simultaneous emergence of RNA and DNA
Nature Chemistry (2019)
-
Emergence of native peptide sequences in prebiotic replication networks
Nature Communications (2017)
-
Opinion: Studies on the origin of life — the end of the beginning
Nature Reviews Chemistry (2017)