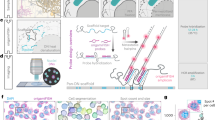
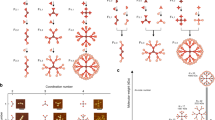
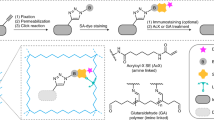
Abstract
The identification and differentiation of a large number of distinct molecular species with high temporal and spatial resolution is a major challenge in biomedical science. Fluorescence microscopy is a powerful tool, but its multiplexing ability is limited by the number of spectrally distinguishable fluorophores. Here, we used (deoxy)ribonucleic acid (DNA)-origami technology to construct submicrometre nanorods that act as fluorescent barcodes. We demonstrate that spatial control over the positioning of fluorophores on the surface of a stiff DNA nanorod can produce 216 distinct barcodes that can be decoded unambiguously using epifluorescence or total internal reflection fluorescence microscopy. Barcodes with higher spatial information density were demonstrated via the construction of super-resolution barcodes with features spaced by ∼40 nm. One species of the barcodes was used to tag yeast surface receptors, which suggests their potential applications as in situ imaging probes for diverse biomolecular and cellular entities in their native environments.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Fournier Bidoz, S. et al. Facile and rapid one-step mass preparation of quantum-dot barcodes. Angew. Chem. Int. Ed. 47, 5577–5581 (2008).
Han, M., Gao, X., Su, J. Z. & Nie, S. Quantum-dot-tagged microbeads for multiplexed optical coding of biomolecules. Nature Biotechnol. 19, 631–635 (2001).
Li, Y., Cu, Y. T. H. & Luo, D. Multiplexed detection of pathogen DNA with DNA-based fluorescence nanobarcodes. Nature Biotechnol. 23, 885–889 (2005).
Marcon, L. et al. ‘On-the-fly’ optical encoding of combinatorial peptide libraries for profiling of protease specificity. Mol. BioSyst. 6, 225–233 (2010).
Xu, H. et al. Multiplexed SNP genotyping using the Qbead system: a quantum dot-encoded microsphere-based assay. Nucleic Acids Res. 31, e43 (2003).
Livet, J. et al. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature 450, 56–62 (2007).
Lin, C., Liu, Y. & Yan, H. Self-assembled combinatorial encoding nanoarrays for multiplexed biosensing. Nano Lett. 7, 507–512 (2007).
Levsky, J. M., Shenoy, S. M., Pezo, R. C. & Singer, R. H. Single-cell gene expression profiling. Science 297, 836–840 (2002).
Braeckmans, K. et al. Encoding microcarriers by spatial selective photobleaching. Nature Mater. 2, 169–173 (2003).
Dejneka, M. J. et al. Rare earth-doped glass microbarcodes. Proc. Natl Acad. Sci. USA 100, 389–393 (2003).
Gudiksen, M. S., Lauhon, L. J., Wang, J., Smith, D. C. & Lieber, C. M. Growth of nanowire superlattice structures for nanoscale photonics and electronics. Nature 415, 617–620 (2002).
Li, X. et al. Controlled fabrication of fluorescent barcode nanorods. ACS Nano 4, 4350–4360 (2010).
Nicewarner-Pena, S. R. Submicrometer metallic barcodes. Science 294, 137–141 (2001).
Pregibon, D. C., Toner, M. & Doyle, P. S. Multifunctional encoded particles for high-throughput biomolecule analysis. Science 315, 1393–1396 (2007).
Geiss, G. K. et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nature Biotechnol. 26, 317–325 (2008).
Xiao, M. et al. Direct determination of haplotypes from single DNA molecules. Nature Methods 6, 199–201 (2009).
Toomre, D. & Bewersdorf, J. A new wave of cellular imaging. Annu. Rev. Cell Dev. Biol. 26, 285–314 (2010).
Seeman, N. C. Nucleic acid junctions and lattices. J. Theor. Biol. 99, 237–247 (1982).
Aldaye, F. A., Palmer, A. L. & Sleiman, H. F. Assembling materials with DNA as the guide. Science 321, 1795–1799 (2008).
Lin, C., Liu, Y. & Yan, H. Designer DNA nanoarchitectures. Biochemistry 48, 1663–1674 (2009).
Nangreave, J., Han, D., Liu, Y. & Yan, H. DNA origami: a history and current perspective. Curr. Opin. Chem. Biol. 14, 608–615 (2010).
Shih, W. M. & Lin, C. Knitting complex weaves with DNA origami. Curr. Opin. Struct. Biol. 20, 276–282 (2010).
Tørring, T., Voigt, N. V., Nangreave, J., Yan, H. & Gothelf, K. V. DNA origami: a quantum leap for self-assembly of complex structures. Chem. Soc. Rev. 40, 5636–5646 (2011).
Rothemund, P. W. K. Folding DNA to create nanoscale shapes and patterns. Nature 440, 297–302 (2006).
Douglas, S. M. et al. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature 459, 414–418 (2009).
Dietz, H., Douglas, S. M. & Shih, W. M. Folding DNA into twisted and curved nanoscale shapes. Science 325, 725–730 (2009).
Andersen, E. S. et al. Self-assembly of a nanoscale DNA box with a controllable lid. Nature 459, 73–76 (2009).
Han, D., Pal, S., Liu, Y. & Yan, H. Folding and cutting DNA into reconfigurable topological nanostructures. Nature Nanotechnol. 5, 712–717 (2010).
Liedl, T., Högberg, B., Tytell, J., Ingber, D. E. & Shih, W. M. Self-assembly of three-dimensional prestressed tensegrity structures from DNA. Nature Nanotechnol. 5, 520–524 (2010).
Han, D. et al. DNA origami with complex curvatures in three-dimensional space. Science 332, 342–346 (2011).
Jungmann, R. et al. Single-molecule kinetics and super-resolution microscopy by fluorescence imaging of transient binding on DNA origami. Nano Lett. 10, 4756–4761 (2010).
Steinhauer, C., Jungmann, R., Sobey, T. L., Simmel, F. C. & Tinnefeld, P. DNA origami as a nanoscopic ruler for super-resolution microscopy. Angew. Chem. Int. Ed. 48, 8870–8873 (2009).
Lund, K. et al. Molecular robots guided by prescriptive landscapes. Nature 465, 206–210 (2010).
Pal, S., Deng, Z., Ding, B., Yan, H. & Liu, Y. DNA-origami-directed self-assembly of discrete silver-nanoparticle architectures. Angew. Chem. Int. Ed. 49, 2700–2704 (2010).
Bui, H. et al. Programmable periodicity of quantum dot arrays with DNA origami nanotubes. Nano Lett. 10, 3367–3372 (2010).
Douglas, S. M., Chou, J. J. & Shih, W. M. DNA–nanotube-induced alignment of membrane proteins for NMR structure determination. Proc. Natl Acad. Sci. USA 104, 6644–6648 (2007).
Hell, S. W. Far-field optical nanoscopy. Science 316, 1153–1158 (2007).
Huang, B., Babcock, H. & Zhuang, X. Breaking the diffraction barrier: super-resolution imaging of cells. Cell 143, 1047–1058 (2010).
Vogelsang, J. et al. Make them blink: probes for super-resolution microscopy. ChemPhysChem. 11, 2475–2490 (2010).
Walter, N. G., Huang, C. Y., Manzo, A. J. & Sobhy, M. A. Do-it-yourself guide: how to use the modern single-molecule toolkit. Nature Methods 5, 475–489 (2008).
Betzig, E. et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science 313, 1642–1645 (2006).
Rust, M. J., Bates, M. & Zhuang, X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nature Methods 3, 793–795 (2006).
Yildiz, A. et al. Myosin V walks hand-over-hand: single fluorophore imaging with 1.5-nm localization. Science 300, 2061–2065 (2003).
Jones, S. A., Shim, S. H., He, J. & Zhuang, X. Fast, three-dimensional super-resolution imaging of live cells. Nature Methods 8, 499–508 (2011).
Gautier, A. et al. An engineered protein tag for multiprotein labeling in living cells. Chem. Biol. 15, 128–136 (2008).
Keppler, A. et al. A general method for the covalent labeling of fusion proteins with small molecules in vivo. Nature Biotechnol. 21, 86–89 (2003).
Klein, T. et al. Live-cell dSTORM with SNAP-tag fusion proteins. Nature Methods 8, 7–9 (2011).
Cunin, F. et al. Biomolecular screening with encoded porous-silicon photonic crystals. Nature Mater. 1, 39–41 (2002).
Matesanz-Isabel, J. et al. New B-cell CD molecules. Immunol. Lett. 134, 104–112 (2011).
Maecker, H. T., McCoy, J. P. & Nussenblatt, R. Standardizing immunophenotyping for the Human Immunology Project. Nature Rev. Immunol. 12, 191–200 (2012).
Acknowledgements
We thank C. Steinhauer and S. P. Laurien for help with super-resolution microscopy software development, the Harvard Center for Biological Imaging, as well as the Nikon Imaging Center at Harvard Medical School, for the use of their microscopes and S. M. Douglas for providing the transmission electron microscopy images used in Supplementary Fig. S5. This work is supported by a National Institutes of Health (NIH) Director's New Innovator Award (1DP2OD007292), a National Science Foundation Faculty Early Career Development Award (CCF1054898), an Office of Naval Research Young Investigator Program Award (N000141110914), an Office of Naval Research grant (N000141010827) and a Wyss Institute for Biologically Engineering Faculty Startup Fund to P.Y., and an NIH Director's New Innovator Award (1DP2OD004641) and a Wyss Institute for Biologically Inspired Engineering Faculty Award to W.M.S. C. Li, D.L. and G.M.C. acknowledge support from the National Human Genome Research Institute, Centers of Excellence in Genomic Science. R.J. acknowledges support from the Alexander von Humboldt-Foundation through a Feodor Lynen fellowship.
Author information
Authors and Affiliations
Contributions
C. Lin conceived the project, designed and conducted the majority of the experiments, analysed the data and prepared the majority of the manuscript. R.J. conceived the super-resolution barcode study, designed and conducted experiments for this study, analysed the data and prepared the manuscript. A.M.L. wrote the MATLAB script for the automated barcode deciphering and prepared the manuscript. C. Li wrote the C script for the barcode geometry characterization. D.L. (with C. Lin) performed the yeast-tagging experiment. G.M.C. championed multiplexed in situ, supervised C. Li and D.L., and critiqued the data and the manuscript. W.M.S. conceived the project, discussed the results and prepared the manuscript. P.Y. conceived, designed and supervised the study, interpreted the data and prepared the manuscript. All authors reviewed and approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information
Supplementary information (PDF 20281 kb)
Supplementary information
High-resolution Fig. S3 (PDF 8483 kb)
Supplementary information
High-resolution Fig. S7 (PDF 5239 kb)
Supplementary information
High-resolution Fig. S8 (PDF 11430 kb)
Supplementary information
High-resolution Fig. S9 (PDF 5958 kb)
Supplementary information
High-resolution Fig. S12 (PDF 5111 kb)
Rights and permissions
About this article
Cite this article
Lin, C., Jungmann, R., Leifer, A. et al. Submicrometre geometrically encoded fluorescent barcodes self-assembled from DNA. Nature Chem 4, 832–839 (2012). https://doi.org/10.1038/nchem.1451
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchem.1451
This article is cited by
-
A Comprehensive Review on the Classification, Uses, Sources of Nanoparticles (NPs) and Their Toxicity on Health
Aerosol Science and Engineering (2023)
-
Single-particle combinatorial multiplexed liposome fusion mediated by DNA
Nature Chemistry (2022)
-
Nanorods with multidimensional optical information beyond the diffraction limit
Nature Communications (2020)
-
Development of a sequencing system for spatial decoding of DNA barcode molecules at single-molecule resolution
Communications Biology (2020)
-
Encoding quantized fluorescence states with fractal DNA frameworks
Nature Communications (2020)