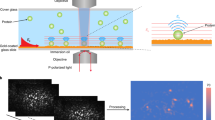
Abstract
Membrane proteins mediate a variety of cellular responses to extracellular signals. Although membrane proteins are studied intensively for their values as disease biomarkers and therapeutic targets, in situ investigation of the binding kinetics of membrane proteins with their ligands has been a challenge. Traditional approaches isolate membrane proteins and then study them ex situ, which does not reflect accurately their native structures and functions. We present a label-free plasmonic microscopy method to map the local binding kinetics of membrane proteins in their native environment. This analytical method can perform simultaneous plasmonic and fluorescence imaging, and thus make it possible to combine the strengths of both label-based and label-free techniques in one system. Using this method, we determined the distribution of membrane proteins on the surface of single cells and the local binding kinetic constants of different membrane proteins. Furthermore, we studied the polarization of the membrane proteins on the cell surface during chemotaxis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Cho, W. H. & Stahelin, R. V. Membrane–protein interactions in cell signaling and membrane trafficking. Annu. Rev. Biophys. Biomol. Struc. 34, 119–151 (2005).
Marinissen, M. J. & Gutkind, J. S. G-protein-coupled receptors and signaling networks: emerging paradigms. Trend. Pharm. Sci. 22, 368–376 (2001).
Hopkins, A. L. & Groom, C. R. The druggable genome. Nature Rev. Drug Disc. 1, 727–730 (2002).
Adams, G. P. & Weiner, L. M. Monoclonal antibody therapy of cancer. Nature Biotech. 23, 1147–1157 (2005).
Rees, D. C., Congreve, M., Murray, C. W. & Carr, R. Fragment-based lead discovery. Nature Rev. Drug Disc. 3, 660–672 (2004).
Salamon, Z., Macleod, H. A. & Tollin, G. Surface plasmon resonance spectroscopy as a tool for investigating the biochemical and biophysical properties of membrane protein systems. 2. Applications to biological systems. Biochim. Biophys. Acta 1331, 131–152 (1997).
Lee, A. G. How lipids affect the activities of integral membrane proteins. Biochim. Biophys. Acta 1666, 62–87 (2004).
Fruh, V., Ijzerman, A. P. & Siegal, G. How to catch a membrane protein in action: a review of functional membrane protein immobilization strategies and their applications. Chem. Rev. 111, 640–656 (2011).
Bally, M. et al. Liposome and lipid bilayer arrays towards biosensing applications. Small 6, 2481–2497 (2010).
Holden, M. A. et al. Direct transfer of membrane proteins from bacteria to planar bilayers for rapid screening by single-channel recording. Nature Chem. Bio. 2, 314–318 (2006).
Dykstra, M. et al. Location is everything: lipid rafts and immune cell signaling. Annu. Rev. Immun. 21, 457–481 (2003).
Sato, T. K., Overduin, M. & Emr, S. D. Location, location, location: membrane targeting directed by PX domains. Science 294, 1881–1885 (2001).
Li, G. Y., Xi, N. & Wang, D. H. Probing membrane proteins using atomic force microscopy. J. Cell. Biochem. 97, 1191–1197 (2006).
Muller, D. J. & Engel, A. Atomic force microscopy and spectroscopy of native membrane proteins. Nature Protocols 2, 2191–2197 (2007).
Groves, J. T., Parthasarathy, R. & Forstner, M. B. Fluorescence imaging of membrane dynamics. Annu. Rev. Biomed. Eng. 10, 311–338 (2008).
Schwarzenbacher, M. et al. Micropatterning for quantitative analysis of protein–protein interactions in living cells. Nature Methods 5, 1053–1060 (2008).
Johnson, A. E. Fluorescence approaches for determining protein conformations, interactions and mechanisms at membranes. Traffic 6, 1078–1092 (2005).
Wallrabe, H. & Periasamy, A. Imaging protein molecules using FRET and FLIM microscopy. Curr. Opin. Biotech. 16, 19–27 (2005).
Axelrod, D. Total internal reflection fluorescence microscopy in cell biology. Traffic 2, 764–774 (2001).
Wang, W. et al. Single cells and intracellular processes studied by a plasmonic-based electrochemical impedance microscopy. Nature Chem. 3, 249–255 (2011).
Huang, B., Yu, F. & Zare, R. N. Surface plasmon resonance imaging using a high numerical aperture microscope objective. Anal. Chem. 79, 2979–2983 (2007).
Kadurin, I. et al. Differential effects of N-glycans on surface expression suggest structural differences between the acid-sensing ion channel (ASIC) 1a and ASIC1b. Biochem. J. 412, 469–475 (2008).
Dell, A. & Morris, H. R. Glycoprotein structure determination mass spectrometry. Science 291, 2351–2356 (2001).
Durand, G. & Seta, N. Protein glycosylation and diseases: blood and urinary oligosaccharides as markers for diagnosis and therapeutic monitoring. Clin. Chem. 46, 795–805 (2000).
Liu S. L. et al. Visualizing the endocytic and exocytic processes of wheat germ agglutinin by quantum dot-based single-particle tracking. Biomaterials 32, 7616–7624 (2011).
Vila-Perello, M., Gallego, R. G. & Andreu, D. A simple approach to well-defined sugar-coated surfaces for interaction studies. ChemBioChem 6, 1831–1838 (2005).
Gingell, D., Todd, I. & Bailey, J. Topography of cell–glass apposition revealed by total internal reflection fluorescence of volume markers. J. Cell Biol. 100, 1334–1338 (1985).
Sato, Y. et al. High mannose-binding lectin with preference for the cluster of 1-2-mannose from the green alga Boodlea coacta is a potent entry inhibitor of HIV-1 and influenza viruses. J. Biol. Chem. 286, 19446–19458 (2011).
Katrlik, J., Skrabana, R., Mislovicova, D. & Gemeiner, P. Binding of D-mannose-containing glycoproteins to D-mannose-specific lectins studied by surface plasmon resonance. Colloid Surf. A 382, 198–202 (2011).
Rathanaswami, P., Babcook, J. & Gallo, M. High-affinity binding measurements of antibodies to cell-surface-expressed antigens. Anal. Biochem. 373, 52–60 (2008).
Troise, F. et al. Differential binding of human immunoagents and herceptin to the ErbB2 receptor. FEBS J. 275, 4967–4979 (2008).
Lehmann, S. et al. An endogenous lectin and one of its neuronal glycoprotein ligands are involved in contact guidance of neuron migration. Proc. Natl Acad. Sci. USA 87, 6455–6459 (1990).
Zieske, J. D., Higashijima, S. C. & Gipson, I. K. Con A-binding and WGA-binding glycoproteins of stationary and migratory corneal epithelium. Invest. Ophthalmol. Vis. Sci. 27, 1205–1210 (1986).
Huppa, J. B. et al. TCR–peptide–MHC interactions in situ show accelerated kinetics and increased affinity. Nature 463, 963–967 (2010).
Kataoka, M. & Tavassoli, M. Polarization of membrane-glycoproteins during monocyte chemotaxis. Exp. Cell Res. 153, 539–543 (1984).
Russo, V. C. et al. Insulin-like growth factor binding protein-2 binding to extracellular matrix plays a critical role in neuroblastoma cell proliferation, migration, and invasion. Endocrinology 146, 4445–4455 (2005).
Albuquerque, E. X., Pereira, E. F. R., Alkondon, M. & Rogers, S. W. Mammalian nicotinic acetylcholine receptors: from structure to function. Physiol. Rev. 89, 73–120 (2009).
Eaton, J. B. et al. Characterization of human alpha 4 beta 2-nicotinic acetylcholine receptors stably and heterologously expressed in native nicotinic receptor-null SH-EP1 human epithelial cells. Mol. Pharmacol. 64, 1283–1294 (2003).
DeFazio-Eli, L. et al. Quantitative assays for the measurement of HER1–HER2 heterodimerization and phosphorylation in cell lines and breast tumors: applications for diagnostics and targeted drug mechanism of action. Breast Cancer Res. 13, R44 (2011).
Manz, B. N. & Groves, J. T. Spatial organization and signal transduction at intercellular junctions. Nature Rev. Mol. Cell Biol. 11, 342–352 (2010).
Pick, H. et al. Monitoring expression and clustering of the ionotropic 5HT3 receptor in plasma membranes of live biological cells. Biochemistry 42, 877–884 (2003).
Acknowledgements
We thank the National Institutes of Health (R21RR026235) for support.
Author information
Authors and Affiliations
Contributions
W.W. and Y.Y. designed and performed the experiments. W.W., Y.Y., S.W., V.J.N. and N.J.T. discussed the results. Q.L. and J.W. provided the cell lines and helped with the immunofluorescence of nAChR. W.W. and N.J.T. wrote the paper. N.J.T. conceived the experiment and supervised the project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information
Supplementary information (PDF 3071 kb)
Supplementary Movie 1
Supplementary Movie 1 (MOV 3660 kb)
Supplementary Movie 2
Supplementary Movie 2 (MOV 2698 kb)
Rights and permissions
About this article
Cite this article
Wang, W., Yang, Y., Wang, S. et al. Label-free measuring and mapping of binding kinetics of membrane proteins in single living cells. Nature Chem 4, 846–853 (2012). https://doi.org/10.1038/nchem.1434
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchem.1434
This article is cited by
-
Effects of Chemical Fixatives on Kinetic Measurements of Biomolecular Interaction on Cell Membrane
The Journal of Membrane Biology (2024)
-
Curvature dependence of BAR protein membrane association and dissociation kinetics
Scientific Reports (2022)
-
Planar photonic chips with tailored angular transmission for high-contrast-imaging devices
Nature Communications (2021)
-
Critical angle reflection imaging for quantification of molecular interactions on glass surface
Nature Communications (2021)
-
Wide-field optical sizing of single nanoparticles with 10 nm accuracy
Science China Physics, Mechanics & Astronomy (2021)