Abstract
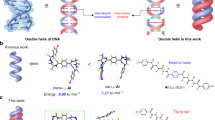
Short nucleotide fragments such as mono- and dinucleotides are generally unable to form stable hydrogen-bonded base pairs or duplexes in water. Within the hydrophobic pockets of enzymes, however, even short fragments form stable duplexes to transmit genetic information. Here, we demonstrate the efficient formation of hydrogen-bonded base pairs from mononucleotides in water through enclathration in the hydrophobic cavities of self-assembled cages. Crystallographic studies and 1H- and 15N-NMR spectroscopy clearly reveals pair-selective recognition of mononucleotides and the selective formation of an anti-Hoogsteen-type base pair in the cage's cavity. Within an analogous expanded cage, dinucleotides are also found to form a stable duplex in water. These results emphasize how hydrogen-bonded base pairing is amplified in a local hydrophobic area isolated from aqueous solution.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Watson, J. D. & Crick, F. H. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature 171, 737–738 (1953).
Saenger, W. Principles of Nucleic Acid Structure (Springer, 1984).
Philp, D. & Stoddart, J. F. Self-assembly in natural and unnatural systems. Angew. Chem. Int. Ed. Engl. 35, 1154–1196 (1996).
Korostelev, A., Trakhanov, S., Laurberg, M. & Noller, H. F. Crystal structure of a 70S ribosome–tRNA complex reveals functional interactions and rearrangements. Cell 126, 1065–1077 (2006).
Westover, K. D., Bushnell, D. A. & Kornberg, R. D. Structural basis of transcription: nucleotide selection by rotation in the RNA polymerase II active center. Cell 119, 481–489 (2004).
Kumazawa, K., Biradha, K., Kusukawa, T., Okano, T. & Fujita, M. Multicomponent assembly of a pyrazine-pillared coordination cage that selectively binds planar guests by intercalation. Angew. Chem. Int. Ed. 42, 3909–3913 (2003).
Hoogsteen, K. The crystal and molecular structure of a hydrogen-bonded complex between 1-methylthymine and 9-methyladenine. Acta Crystallogr. 16, 907–916 (1963).
Hosseini, M. W., Blacker, A. J. & Lehn, J.-M. Multiple molecular recognition and catalysis. A multifunctional anion receptor bearing an anion binding site, an intercalating group, and a catalytic site for nucleotide binding and hydrolysis. J. Am. Chem. Soc. 112, 3896–3904 (1990).
Kobayashi, K., Asakawa, Y., Kato, Y. & Aoyama, Y. Complexation of hydrophobic sugars and nucleosides in water with tetrasulfonate derivatives of resorcinol cyclic tetramer having a polyhydroxy aromatic cavity: importance of guest–host CH–π interaction. J. Am. Chem. Soc. 114, 10307–10313 (1992).
Jasper, C., Schrader, T., Panitzky, J. & Klärner, F.-G. Selective complexation of N-alkylpyridinium salts: recognition of NAD in water . Angew. Chem. Int. Ed. 41, 1355–1358 (2002).
Butterfield, S. M. & Waters, M. L. A designed β-hairpin peptide for molecular recognition of ATP in water . J. Am. Chem. Soc. 125, 9580–9581 (2003).
McCleskey, S. C., Griffin, M. J., Schneider, S. E., McDevitt, J. T. & Anslyn, E. V. Differential receptors create patterns diagnostic for ATP and GTP. J. Am. Chem. Soc. 125, 1114–1115 (2003).
Prins, L. J., Reinhoudt, D. N. & Timmerman, P. Noncovalent synthesis using hydrogen bonding. Angew. Chem. Int. Ed. 40, 2382–2426 (2001).
Nowick, J. S., Chen, J. S. & Noronha, G. Molecular recognition in micelles: the roles of hydrogen bonding and hydrophobicity in adenine–thymine base-pairing in SDS micelles. J. Am. Chem. Soc. 115, 7636–7644 (1993).
Kato, Y., Conn, M. M. & Rebek, J. Jr Hydrogen bonding in water using synthetic receptors. Proc. Natl Acad. Sci. USA 92, 1208–1212 (1995).
Asanuma, H., Ban, T., Gotoh, S., Hishiya, T. & Komiyama, M. Hydrogen bonding in water by poly(vinyldiaminotriazine) for the molecular recognition of nucleic acid bases and their derivatives. Macromolecules 31, 371–377 (1998).
Hirschberg, J. H. K. K. et al. Helical self-assembled polymers from cooperative stacking of hydrogen-bonded pairs. Nature 407, 167–170 (2000).
Raszka, M. & Kaplan, N. O. Association by hydrogen bonding of mononucleotides in aqueous solution. Proc. Natl Acad. Sci. USA 69, 2025–2029 (1972).
Yoshizawa, M. et al. Discrete stacking of large aromatic molecules within organic-pillared coordination cages. Angew. Chem. Int. Ed. 44, 1810–1813 (2005).
Cheng, D. M. & Sarma, R. H. Intimate details of the conformational characteristics of deoxyribonucleoside monophosphates in aqueous solution. J. Am. Chem. Soc. 99, 7333–7348 (1977).
Hosur, R. V. & Govil, G. Sequence effects in structures of the dinucleotides d-pApT and d-pTpA. J. Mol. Struct. 72, 261–267 (1981).
Acknowledgements
We thank Masaki Kawano of The University of Tokyo for help with X-ray crystallographic analysis.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Information
Supplementary information (PDF 2166 kb)
Supplementary Information
Crystallographic data for homoduplex of compound 9 inside cage 4 (CIF 60 kb)
Supplementary Information
Crystallographic data for homoduplex of compound 13 inside cage 3 (CIF 24 kb)
Rights and permissions
About this article
Cite this article
Sawada, T., Yoshizawa, M., Sato, S. et al. Minimal nucleotide duplex formation in water through enclathration in self-assembled hosts. Nature Chem 1, 53–56 (2009). https://doi.org/10.1038/nchem.100
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchem.100
This article is cited by
-
A host–guest semibiological photosynthesis system coupling artificial and natural enzymes for solar alcohol splitting
Nature Communications (2021)
-
Chemical reactivity under nanoconfinement
Nature Nanotechnology (2020)
-
Strategies for binding multiple guests in metal–organic cages
Nature Reviews Chemistry (2019)
-
Polyaromatic molecular peanuts
Nature Communications (2017)
-
Iterative design of a helically folded aromatic oligoamide sequence for the selective encapsulation of fructose
Nature Chemistry (2015)