Abstract
The emergence of resistance to poly-ADP-ribose polymerase inhibitors (PARPi) poses a threat to the treatment of BRCA1 and BRCA2 (BRCA1/2)-deficient tumours1. Stabilization of stalled DNA replication forks is a recently identified PARPi-resistance mechanism that promotes genomic stability in BRCA1/2-deficient cancers2. Dissecting the molecular pathways controlling genomic stability at stalled forks is critical. Here we show that EZH2 localizes at stalled forks where it methylates Lys27 on histone 3 (H3K27me3), mediating recruitment of the MUS81 nuclease. Low EZH2 levels reduce H3K27 methylation, prevent MUS81 recruitment at stalled forks and cause fork stabilization. As a consequence, loss of function of the EZH2/MUS81 axis promotes PARPi resistance in BRCA2-deficient cells. Accordingly, low EZH2 or MUS81 expression levels predict chemoresistance and poor outcome in patients with BRCA2-mutated tumours. Moreover, inhibition of Ezh2 in a murine Brca2−/− breast tumour model is associated with acquired PARPi resistance. Our findings identify EZH2 as a critical regulator of genomic stability at stalled forks that couples histone modifications to nuclease recruitment. Our data identify EZH2 expression as a biomarker of BRCA2-deficient tumour response to chemotherapy.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Lord, C. J. & Ashworth, A. Mechanisms of resistance to therapies targeting BRCA-mutant cancers. Nat. Med. 19, 1381–1388 (2013).
Ray Chaudhuri, A. et al. Replication fork stability confers chemoresistance in BRCA-deficient cells. Nature 535, 382–387 (2016).
Farmer, H. et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434, 917–921 (2005).
Konstantinopoulos, P. A., Ceccaldi, R., Shapiro, G. I. & D’Andrea, A. D. Homologous recombination deficiency: exploiting the fundamental vulnerability of ovarian cancer. Cancer Discov. 5, 1137–1154 (2015).
Moynahan, M. E., Cui, T. Y. & Jasin, M. Homology-directed DNA repair, mitomycin-C resistance, and chromosome stability is restored with correction of a Brca1 mutation. Cancer Res. 61, 4842–4850 (2001).
Schlacher, K. et al. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell 145, 529–542 (2011).
Bouwman, P. et al. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat. Struct. Mol. Biol. 17, 688–695 (2010).
Alabert, C. & Groth, A. Chromatin replication and epigenome maintenance. Nat. Rev. Mol. Cell Biol. 13, 153–167 (2012).
Clemente-Ruiz, M. & Prado, F. Chromatin assembly controls replication fork stability. EMBO Rep. 10, 790–796 (2009).
Göllner, S. et al. Loss of the histone methyltransferase EZH2 induces resistance to multiple drugs in acute myeloid leukemia. Nat. Med. 23, 69–78 (2016).
de Vries, N. A. et al. Prolonged Ezh2 depletion in glioblastoma causes a robust switch in cell fate resulting in tumor progression. Cell Rep. 10, 383–397 (2015).
Morey, L. & Helin, K. Polycomb group protein-mediated repression of transcription. Trends Biochem. Sci. 35, 323–332 (2010).
Bracken, A. P. et al. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J. 22, 5323–5335 (2003).
Campbell, S., Ismail, I. H., Young, L. C., Poirier, G. G. & Hendzel, M. J. Polycomb repressive complex 2 contributes to DNA double-strand break repair. Cell Cycle 12, 2675–2683 (2013).
Chou, D. M. et al. A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc. Natl Acad. Sci. USA 107, 18475–18480 (2010).
Hansen, K. H. et al. A model for transmission of the H3K27me3 epigenetic mark. Nat. Cell Biol. 10, 1291–300 (2008).
Ceccaldi, R. et al. Homologous-recombination-deficient tumours are dependent on Polh-mediated repair. Nature 518, 258–262 (2015).
Kais, Z. et al. FANCD2 maintains fork stability in BRCA1/2-deficient tumors and promotes alternative end-joining DNA repair. Cell Rep. 15, 2488–2499 (2016).
Cancer, T. & Atlas, G. Integrated genomic analyses of ovarian carcinoma. Nature 474, 609–615 (2011).
McCabe, M. T. et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature 492, 108–112 (2012).
Kim, K. H. & Roberts, C. W. M. Targeting EZH2 in cancer. Nat. Med. 22, 128–134 (2016).
Branzei, D. & Foiani, M. Maintaining genome stability at the replication fork. Nat. Rev. Mol. Cell Biol. 11, 208–219 (2010).
Alabert, C. et al. Nascent chromatin capture proteomics determines chromatin dynamics during DNA replication and identifies unknown fork components. Nat. Cell Biol. 16, 281–293 (2014).
Piunti, A. et al. Polycomb proteins control proliferation and transformation independently of cell cycle checkpoints by regulating DNA replication. Nat. Commun. 5, 3649 (2014).
Alabert, C. et al. Two distinct modes for propagation of histone PTMs across the cell cycle. Genes Dev. 29, 585–590 (2015).
Thomas Leung, K. H., El Hassan, M. A. & Bremner, R. A rapid and efficient method to purify proteins at replication forks under native conditions. Biotechniques 55, 204–206 (2013).
Vassin, V. M., Anantha, R. W., Sokolova, E., Kanner, S. & Borowiec, J. A. Human RPA phosphorylation by ATR stimulates DNA synthesis and prevents ssDNA accumulation during DNA-replication stress. J. Cell Sci. 122, 4070–4080 (2009).
Petermann, E. & Helleday, T. Pathways of mammalian replication fork restart. Nat. Rev. Mol. Cell Biol. 11, 683–687 (2010).
Cortez, D. Preventing replication fork collapse to maintain genome integrity. DNA Repair 32, 149–157 (2015).
Petermann, E., Orta, M. L., Issaeva, N., Schultz, N. & Helleday, T. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol. Cell 37, 492–502 (2010).
Mirzoeva, O. K. & Petrini, J. H. J. DNA replication-dependent nuclear dynamics of the Mre11 complex. Mol. Cancer Res. 1, 207–218 (2003).
Jonkers, J. et al. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat. Genet. 29, 418–425 (2001).
Schwartzentruber, J., Korshunov, A. & Liu, X. Y. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 482, 226–231 (2012).
Hanada, K. et al. The structure-specific endonuclease Mus81 contributes to replication restart by generating double-strand DNA breaks. Nat. Struct. Mol. Biol. 14, 1096–1104 (2007).
Chen, H. et al. Sae2 promotes DNA damage resistance by removing the Mre11–Rad50–Xrs2 complex from DNA and attenuating Rad53 signaling. Proc. Natl Acad. Sci. USA 112, E1880–E1887 (2015).
The Cancer Genome Atlas NetworkComprehensive molecular portraits of human breast tumours. Nature 490, 61–70 (2012).
Rottenberg, S. et al. Selective induction of chemotherapy resistance of mammary tumors in a conditional mouse model for hereditary breast cancer. Proc. Natl Acad. Sci. USA 104, 12117–12122 (2007).
Thangavel, S. et al. DNA2 drives processing and restart of reversed replication forks in human cells. J. Cell Biol. 208, 545–562 (2015).
Gong, Z., Cho, Y.-W., Kim, J.-E., Ge, K. & Chen, J. Accumulation of Pax2 transactivation domain interaction protein (PTIP) at sites of DNA breaks via RNF8-dependent pathway is required for cell survival after DNA damage. J. Biol. Chem. 284, 7284–7293 (2009).
The Cancer Genome Atlas NetworkIntegrated genomic characterization of endometrial carcinoma. Nature 497, 67–73 (2013).
Acknowledgements
We thank all the members of the D’Andrea laboratory for their useful comments, suggestions and help. We thank D. Chowdhury and G. I. Shapiro for helpful discussions on the project and their laboratories for help and reagents (the Shapiro laboratory also provided MDA-MB-436 EV and HA-BRCA1). We thank K. Mouw for helpful discussions and critical reading of the manuscript. We thank S. Orkin (DFCI, Boston, USA) and M. I. Aladjem (NCI, NIH, Bethesda, MD) for providing us with the siEZH2-resistant WT, ΔHMT EZH2 cDNA and with full-length MUS81 plasmids, respectively. We thank the people from the Preclinical Intervention Unit of the Mouse Clinic for Cancer and Ageing (MCCA) at the NKI for their technical support with the animal studies and J. R. de Ruiter for help with RNA-Seq analysis of the KB2P tumour panel. We thank P. Germain for a preliminary analysis on ovarian carcinoma TCGA gene expression data. B.R. is a recipient of an Italian Association for Cancer Research (AIRC) Fellowship for Abroad and an Ovarian Cancer Research Fellowship (OCRF) Ann Schreiber Mentored Investigator Award. This research was supported by a Stand Up To Cancer-Ovarian Cancer Research Fund Alliance-National Ovarian Cancer Coalition Dream Team Translational Research Grant (Grant Number: SU2C-AACR-DT16-15). Stand Up To Cancer is a programme of the Entertainment Industry Foundation. Research grants are administered by the American Association for Cancer Research, the scientific partner of SU2C. This work was also supported by grants from the US National Institutes of Health (R01DK43889, R01HL052725, P01HL048546, P50CA168504), the United States Department of Defense (BC151331P1), the Breast Cancer Research Foundation, and the Ludwig Center at Harvard (to A.D.D.), the Dutch Cancer Society (2014-6532 to S.R. and J.J.), the Swiss National Science Foundation (310030_156869 to S.R.), and the European Union (ERC CoG-681572 to S.R.).
Author information
Authors and Affiliations
Contributions
B.R. conceived the study, performed experiments, analysed data and wrote the manuscript. E.G. and A.A.D. performed and analysed the animal, gene expression and IHC experiments under the supervision of M.v.d.V., J.J. and S.R. H.Y. and R.v.d.S. performed experiments, A.D.D. and M.v.d.V. contributed to mouse work, P.A.K. curated TCGA data sets for Fig. 1g and Supplementary Fig. 5d and provided clinical perspectives. R.C. performed bioinformatic analysis on TCGA gene expression, reviewed data and provided expertise, S.R. reviewed in vivo and IHC data and provided expertise, and A.D.D. conceived the study and wrote the manuscript. All authors approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 EZH2-mediated drug resistance depends on its methyltransferase activity and extends to cisplatin.
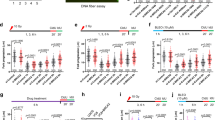
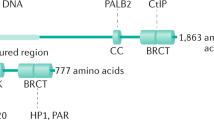
(a) EZH2 expression in OvCa TCGA (n = 494 OvCa (grade 1, n = 5; grade 2, n = 61; grade 3, n = 428) and n = 8 control samples). Each dot represents a Z-score for a tumor. Error bars represent s.e.m. and the black bar represents the median. P = 0.00063. (b) MKi67 gene expression in ovarian (grade 2/3 serous n = 143 and grade 1 n = 20; ∗∗∗P = 0.00034), breast (basal-like n = 80 and rest n = 421; ∗∗∗P = 0.00045) and uterine (serous-like n = 60 compared to the rest n = 172; ∗∗∗P = 0.00067) cancers. Expression values are represented as the mean ± s.e.m. of Z-scores. c,d, EZH2 expression in breast carcinoma TCGA (c) and in ovarian TCGA (d). (e) EZH2 expression in breast TCGA (n = 501. samples). Bars represent mean ± s.e.m. and ∗∗∗P = 0.000136. (f–l) Clonogenic assays in A2780 (f), KURAMOCHI (g) or in HCC1937 (h), MDA-MB-436 (i), SUM149 or UWB1.289 (l). m, Immunoblot of one out of 3 representative experiment of HeLa cells treated with vehicle (DMSO) or GSK126. (n,o) Clonogenic assay in HeLa (n), A2780 (o) treated with DMSO or GSK126 under cisplatin. (p) EZH2 domains with the F672I/H694A/R732K mutations of ΔHMT mutant. (m) Clonogenic assays in EV, WT or ΔHMT VU423 cells treated with rucaparib. (q,r) Chromosome breakage analysis of VU423 (q) and KURAMOCHI cells (r) treated with DMSO or GSK126 and with rucaparib. For a,b, significance was assessed with one-way Anova. Correlation was assessed with Pearson test r = 0.68, P = 0.00026 (c) and r = 0.46, P = 0.00067 (d). Data in f–i represent mean ± s.e.m. of n = 3 independent experiments. Data in n–r represent mean ± s.e.m. of n = 3 independent experiments. n: ∗P = 0.016, o: ∗P = 0.028, p: ∗∗P = 0.00045, q: ∗∗P = 0.0021 and r: from left, ∗∗∗P = 0.00011 and ∗P = 0.038 Significance in e,n–r was assessed with the two-sided Student’s t test. In b,e,n–r, ∗P < 0.05, ∗∗P < 0.01 and ∗∗∗P < 0.001, NS = non significant with exact P values (with a confidence interval of 95%) noted in legends for relevant panels. Unprocessed original scans of blots are shown in Supplementary Fig. 6.
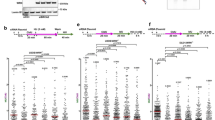
Supplementary Figure 2 Fork protection and not HR restoration is associated with EZH2 loss at stalled forks.
(a) DR-GFP assay in cells transfected with siRNAs and/or treated with GSK126. (b,c) Quantification of RAD51 foci in HeLa (b) and VU423 (c) cells treated as indicated and subjected to HU treatment. The number of RAD51 positive cells is expressed as fold change of siScr-transfected sample, whose value is set to 1. (d) Schematic for fork protection experiments. Box and whiskers plot show CldU to IdU ratio. P = 0.001. (e) Fork protection experiment and immunoblot analysis of VU423 cells stably expressing empty vector (EV) or Chromosome 13 (containing BRCA2 gene) and treated with DMSO or GSK126. P = 0.04. (f) Fork protection experiment and immunoblot analysis of MDA436 cells stably expressing an empty vector (EV) or a vector expressing HA-BRCA1 and treated with DMSO or GSK126. P = NS, not significant. (g) Schematic for the labeling of cells with IdU and CldU for fork protection experiments. Box plot show CldU to IdU ratio. P = 0.0002. (h) PLA assays in VU423 cells transfected with the indicated siRNAs and co-stained with antibodies against pSer4/8 RPA and EZH2. Bars indicate the number of cells with more than 10 PLA foci. n = 100 cells were counted per condition. Data in a,h represent mean ± s.e.m. of n = 3 independent experiments. Significance was assessed with the χ2 significance test for trend in proportions. a: ∗∗P = 0.0049, h: ∗∗P = 0.0064. Data in b,c represent mean ± s.e.m. of n = 3 independent experiments. Significance was assessed with two-sided Student’s t test. ∗P = 0.021. Box plots in d–f and g display median bar, first-third quartile box and min-to-max value whiskers. Analysis was performed by measuring analogue tracts on n = 100 fibers per condition on n = 3 independent experiments. Significance of difference among all the groups was assessed with the one-sided Kruskal-Wallis test. In a–c and h, ∗P < 0.05, ∗∗P < 0.01, NS = non significant with exact P values (with a confidence interval of 95%) noted in legends for relevant panels. Unprocessed original scans of blots are shown in Supplementary Fig. 6.
Supplementary Figure 3 EZH2 mediates fork degradation by specifically recruiting the endonuclease MUS81 through H3K27 methylation.
(a) Schematic of fork protection experiments with CldU and IdU incorporation for VU423 cells. Box plots for CldU to IdU ratio of VU423 cells transduced with EV, WT or ΔHMT are shown. P = 0.0006. (b) One representative Immunoblot analysis of three independently performed and with similar results of VU423 cell transduced with EV or WT EZH2 and transfected with the indicated siRNAs. (c) Schematic for the labeling with IdU and CldU of cells expressing EV or WT EZH2 for fork protection experiments. Box plots show CldU/IdU ratio of cells expressing the indicated cDNA and transfected with nuclease siRNAs (MUS81_5 was used). P = 0.000012. (d) Pull-down in 293T whole cell lysate with unmethylated or trimethylated histone H3 at lysine 27 (H3 or H3K27me3) depicting differential MUS81 but not MRE11 pull-down. EED binding to H3K27me3 is used as a positive control. (e,f) Pull-down with trimethylated histone H3 at lysine 27 (H3K27me3) in 293T whole cell lysate expressing full-length or truncated MUS81 (e) or after in vitro transcription and translation of the MUS81 constructs (TnT, f). EED binding to H3K27me3 is used as a positive control. (g) Pull-down after in vitro transcription and translation of the MUS81 constructs (TnT) performed with dimethylated histone H3 at lysine 27 (H3K27me2). Box plots in a and c display median bar, first-third quartile box and min-to-max value whiskers. Analysis was performed by measuring analogue tracts on n = 100 fibers per condition on n = 3 independent experiments. Significance of difference among all the groups was assessed with the one-sided Kruskal-Wallis test. Data in e–h represent one out of 3 independent experiments with similar results. Unprocessed original scans of blots are shown in Supplementary Fig. 6.
Supplementary Figure 4 MUS81 levels are not dependent on EZH2-mediated transcriptional activity and separate BRCA2-mutated ovarian carcinoma patients based on their PFS after chemotherapy.
(a) aniPOND of one out of 3 independent experiments with similar results were performed in 293T cells transduced with control (shScr) or EZH2 specific shRNAs (EZH2_1 and EZH2_4) showing recruitment of proteins at replicating (untreated, UT) and stalled (HU treated) forks. (b) Immunofluorescence analysis of one out of 3 independent experiments with similar results of colocalization of MRE11 and MUS81 with sites of stalled replication, as marked by CldU incorporation upon HU treatment. Scale bar, 1 μm. (c) Immunoblot analysis of one out of 3 independent experiments with similar results of VU423 and HeLa cells transfected with the indicated siRNAs. (d) Progression-free survival (PFS) after platinum chemotherapy of TCGA patients harboring BRCA wild-type (n = 82 samples), BRCA1-mutated (n = 27 samples) or BRCA2-mutated (n = 34 samples) ovarian carcinomas. Statistical significance was assessed by the two-sided log-rank test, P = 0.055 with a 95% confidence interval. Unprocessed original scans of blots are shown in Supplementary Fig. 6.
Supplementary Figure 5 Ezh2 inhibition shortens the overall survival of mice harboring KB2P-derived tumors without affecting tumor proliferation.
(a) Outline of the PARPi in vivo intervention study. A spontaneous Brca2-/p53-deficient tumor was generated and re-transplanted into syngenic WT mice. When the tumors reached 200 mm3, they were treated with either vehicle (n = 9), GSK126 (n = 9), olaparib (n = 9) or olaparib + GSK126 (n = 8). (b) Overall survival of mice orthotopically injected with KB2P-derived tumors. (c) Tumor growth curve of tumors treated with vehicle (grey) or GSK126 (blue). Data are represented as mean ± s.d. In b, the two-sided log-rank (Mantel-Cox) P value is indicated, with a confidence interval of 95%. In c, significance was assessed by the two-sided Student’s t test, ns = non significant.
Supplementary information
Supplementary Information
Supplementary Information (PDF 12120 kb)
Supplementary Table 1
Supplementary Information (XLSX 35 kb)
Rights and permissions
About this article
Cite this article
Rondinelli, B., Gogola, E., Yücel, H. et al. EZH2 promotes degradation of stalled replication forks by recruiting MUS81 through histone H3 trimethylation. Nat Cell Biol 19, 1371–1378 (2017). https://doi.org/10.1038/ncb3626
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb3626
This article is cited by
-
Drug resistance in ovarian cancer: from mechanism to clinical trial
Molecular Cancer (2024)
-
PARP inhibitors: enhancing efficacy through rational combinations
British Journal of Cancer (2023)
-
Leveraging the replication stress response to optimize cancer therapy
Nature Reviews Cancer (2023)
-
The chromatin network helps prevent cancer-associated mutagenesis at transcription-replication conflicts
Nature Communications (2023)
-
Treatment of Ovarian Cancer Beyond PARP Inhibition: Current and Future Options
Drugs (2023)