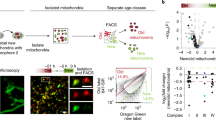
Abstract
Most differentiated cells convert glucose to pyruvate in the cytosol through glycolysis, followed by pyruvate oxidation in the mitochondria. These processes are linked by the mitochondrial pyruvate carrier (MPC), which is required for efficient mitochondrial pyruvate uptake. In contrast, proliferative cells, including many cancer and stem cells, perform glycolysis robustly but limit fractional mitochondrial pyruvate oxidation. We sought to understand the role this transition from glycolysis to pyruvate oxidation plays in stem cell maintenance and differentiation. Loss of the MPC in Lgr5-EGFP-positive stem cells, or treatment of intestinal organoids with an MPC inhibitor, increases proliferation and expands the stem cell compartment. Similarly, genetic deletion of the MPC in Drosophila intestinal stem cells also increases proliferation, whereas MPC overexpression suppresses stem cell proliferation. These data demonstrate that limiting mitochondrial pyruvate metabolism is necessary and sufficient to maintain the proliferation of intestinal stem cells.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Warburg, O. On respiratory impairment in cancer cells. Science 124, 269–270 (1956).
Birsoy, K. et al. An essential role of the mitochondrial electron transport chain in cell proliferation is to enable aspartate synthesis. Cell 162, 540–551 (2015).
Sullivan, L. B. et al. Supporting aspartate biosynthesis is an essential function of respiration in proliferating cells. Cell 162, 552–563 (2015).
Flavell, R. B. Mitochondrion as a multifunctional organelle. Nature 230, 504–506 (1971).
Martínez-Reyes, I. et al. TCA cycle and mitochondrial membrane potential are necessary for diverse biological functions. Mol. Cell 61, 199–209 (2016).
Simsek, T. et al. The distinct metabolic profile of hematopoietic stem cells reflects their location in a hypoxic niche. Cell Stem Cell 7, 380–390 (2010).
Ito, K. & Suda, T. Metabolic requirements for the maintenance of self-renewing stem cells. Nat. Rev. Mol. Cell Biol. 15, 243–256 (2014).
Stringari, C. et al. Metabolic trajectory of cellular differentiation in small intestine by Phasor Fluorescence Lifetime Microscopy of NADH. Sci. Rep. 2, 568 (2012).
Fan, Y.-Y. et al. A bioassay to measure energy metabolism in mouse colonic crypts, organoids, and sorted stem cells. Am. J. Physiol. Gastrointest. Liver Physiol. 309, G1–G9 (2015).
Rodríguez-Colman, M. J. et al. Interplay between metabolic identities in the intestinal crypt supports stem cell function. Nature 543, 424–427 (2017).
Berger, E. et al. Mitochondrial function controls intestinal epithelial stemness and proliferation. Nat. Commun. 7, 13171 (2016).
Bricker, D. K. et al. A mitochondrial pyruvate carrier required for pyruvate uptake in yeast, Drosophila, and humans. Science 337, 96–100 (2012).
Herzig, S. et al. Identification and functional expression of the mitochondrial pyruvate carrier. Science 337, 93–96 (2012).
Schell, J. C. et al. A role for the mitochondrial pyruvate carrier as a repressor of the Warburg effect and colon cancer cell growth. Mol. Cell 56, 400–413 (2014).
Sato, T. et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265 (2009).
Sato, T. & Clevers, H. Primary mouse small intestinal epithelial cell cultures. Methods Mol. Biol. 945, 319–328 (2013).
Uhlen, M. et al. Towards a knowledge-based Human Protein Atlas. Nat. Biotechnol. 28, 1248–1250 (2010).
Uhlen, M., Pontén, F. & Lindskog, C. Charting the human proteome: understanding disease using a tissue-based atlas. Science 347, 1274–1274 (2015).
Jiang, H. & Edgar, B. A. Intestinal stem cell function in Drosophila and mice. Curr. Opin. Genet. Dev. 22, 354–360 (2012).
Li, H. & Jasper, H. Gastrointestinal stem cells in health and disease: from flies to humans. Dis. Model Mech. 9, 487–499 (2016).
Dutta, D. et al. Regional cell-specific transcriptome mapping reveals regulatory complexity in the adult Drosophila midgut. Cell Rep. 12, 346–358 (2015).
Buchon, N. et al. Morphological and molecular characterization of adult midgut compartmentalization in Drosophila. Cell Rep. 3, 1725–1738 (2013).
Wu, J. S. & Luo, L. A protocol for mosaic analysis with a repressible cell marker (MARCM) in Drosophila. Nat. Protoc. 1, 2583–2589 (2006).
Micchelli, C. A. & Perrimon, N. Evidence that stem cells reside in the adult Drosophila midgut epithelium. Nature 439, 475–479 (2006).
Ohlstein, B. & Spradling, A. The adult Drosophila posterior midgut is maintained by pluripotent stem cells. Nature 439, 470–474 (2006).
Li, H. et al. Drosophila larvae synthesize the putative oncometabolite L-2-hydroxyglutarate during normal developmental growth. Proc. Natl Acad. Sci. USA 114, 1353–1358 (2017).
Micchelli, C. A. Whole-mount immunostaining of the adult Drosophila gastrointestinal tract. Methods 68, 273–279 (2014).
Vigueira, P. A. et al. Mitochondrial pyruvate carrier 2 hypomorphism in mice leads to defects in glucose-stimulated insulin secretion. Cell Rep. 7, 2042–2053 (2014).
Vacanti, N. M. et al. Regulation of substrate utilization by the mitochondrial pyruvate carrier. Mol. Cell 56, 425–435 (2014).
Bender, T., Pena, G. & Martinou, J.-C. Regulation of mitochondrial pyruvate uptake by alternative pyruvate carrier complexes. EMBO J. 34, 911–924 (2015).
Yang, C. et al. Glutamine oxidation maintains the TCA cycle and cell survival during impaired mitochondrial pyruvate transport. Mol. Cell 56, 414–424 (2014).
Yin, X. et al. Niche-independent high-purity cultures of Lgr5 + intestinal stem cells and their progeny. Nat. Methods 11, 106–112 (2014).
Aires, C. C. P. et al. Pyruvate uptake is inhibited by valproic acid and metabolites in mitochondrial membranes. FEBS Lett. 582, 3359–3366 (2008).
Benavides, J., Martin, A., Ugarte, M. & Valdivieso, F. Inhibition by valproic acid of pyruvate uptake by brain mitochondria. Biochem. Pharmacol. 31, 1633–1636 (1982).
Dailey, M. J. Nutrient-induced intestinal adaption and its effect in obesity. Physiol. Behav. 136, 74–78 (2014).
Beyaz, S. et al. High-fat diet enhances stemness and tumorigenicity of intestinal progenitors. Nature 531, 53–58 (2016).
Cao, F. et al. Transcriptional and functional profiling of human embryonic stem cell-derived cardiomyocytes. PLoS ONE 3, e3474 (2008).
Chen, B.-Z. et al. Identification of microRNAs expressed highly in pancreatic islet-like cell clusters differentiated from human embryonic stem cells. Cell Biol. Int. 35, 29–37 (2011).
Ralston, A. et al. Gata3 regulates trophoblast development downstream of Tead4 and in parallel to Cdx2. Development 137, 395–403 (2010).
Muntean, A. G. et al. The PAF complex synergizes with MLL fusion proteins at HOX loci to promote leukemogenesis. Cancer Cell 17, 609–621 (2010).
Keller, M. A. et al. Transcriptional regulatory network analysis of developing human erythroid progenitors reveals patterns of coregulation and potential transcriptional regulators. Physiol. Genomics 28, 114–128 (2006).
Tateno, H. et al. Glycome diagnosis of human induced pluripotent stem cells using lectin microarray. J. Biol. Chem. 286, 20345–20353 (2011).
Nishino, K. et al. DNA methylation dynamics in human induced pluripotent stem cells over time. PLoS Genet. 7, e1002085 (2011).
Saito, S. et al. Possible linkages between the inner and outer cellular states of human induced pluripotent stem cells. BMC Syst. Biol. 5 (Suppl. 1), S17 (2011).
Wang, X.-M. et al. The gene expression profiles of induced pluripotent stem cells from individuals with childhood cerebral adrenoleukodystrophy are consistent with proposed mechanisms of pathogenesis. Stem Cell Res. Ther. 3, 39 (2012).
Flores, A. et al. Lactate dehydrogenase activity drives hair follicle stem cell activation. Nat. Cell Biol. http://dx.doi.org/10.1038/ncb3575 (2017).
Camarda, R. et al. Inhibition of fatty acid oxidation as a therapy for MYC-overexpressing triple-negative breast cancer. Nat. Med. 22, 427–432 (2016).
Boroughs, L. K. & DeBerardinis, R. J. Metabolic pathways promoting cancer cell survival and growth. Nat. Cell Biol. 17, 351–359 (2015).
Edmunds, L. R. et al. c-Myc programs fatty acid metabolism and dictates acetyl-CoA abundance and fate. J. Biol. Chem. 289, 25382–25392 (2014).
Pate, K. T. et al. Wnt signaling directs a metabolic program of glycolysis and angiogenesis in colon cancer. EMBO J. 33, 1454–1473 (2014).
Gerstein, M. B. et al. Architecture of the human regulatory network derived from ENCODE data. Nature 489, 91–100 (2012).
Wang, J. et al. Factorbook.org: a Wiki-based database for transcription factor-binding data generated by the ENCODE consortium. Nucleic Acids Res. 41, D171–D176 (2013).
Wang, J. et al. Sequence features and chromatin structure around the genomic regions bound by 119 human transcription factors. Genome Res. 22, 1798–1812 (2012).
Karolchik, D. et al. The UCSC Genome Browser database: 2014 update. Nucleic Acids Res. 42, D764–D770 (2014).
McCool, K. W., Xu, X., Singer, D. B., Murdoch, F. E. & Fritsch, M. K. The role of histone acetylation in regulating early gene expression patterns during early embryonic stem cell differentiation. J. Biol. Chem. 282, 6696–6706 (2007).
Roostaee, A., Benoit, Y. D., Boudjadi, S. & Beaulieu, J.-F. Epigenetics in intestinal epithelial cell renewal. J. Cell. Physiol. 231, 2361–2367 (2016).
Golob, J. L., Paige, S. L., Muskheli, V., Pabon, L. & Murry, C. E. Chromatin remodeling during mouse and human embryonic stem cell differentiation. Dev. Dyn. 237, 1389–1398 (2008).
Barker, N. et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–1007 (2007).
Gray, L. R. et al. Hepatic mitochondrial pyruvate carrier 1 is required for efficient regulation of gluconeogenesis and whole-body glucose homeostasis. Cell Metab. 22, 669–681 (2015).
Moolenbeek, C. & Ruitenberg, E. J. The ‘Swiss roll’: a simple technique for histological studies of the rodent intestine. Lab. Anim. 15, 57–59 (1981).
Daniels, R. W., Rossano, A. J., Macleod, G. T. & Ganetzky, B. Expression of multiple transgenes from a single construct using viral 2A peptides in Drosophila. PLoS ONE 9, e100637 (2014).
Sato, T. et al. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature 469, 415–418 (2011).
Ootani, A. et al. Sustained in vitro intestinal epithelial culture within a Wnt-dependent stem cell niche. Nat. Med. 15, 701–706 (2009).
Simmini, S. et al. Transformation of intestinal stem cells into gastric stem cells on loss of transcription factor Cdx2. Nat. Commun. 5, 5728 (2014).
Weber, K., Mock, U., Petrowitz, B., Bartsch, U. & Fehse, B. Lentiviral gene ontology (LeGO) vectors equipped with novel drug-selectable fluorescent proteins: new building blocks for cell marking and multi-gene analysis. Gene Ther. 17, 511–520 (2010).
Acknowledgements
We thank B. Edgar (University of Utah, USA) for stocks and reagents, C. Micchelli (Washington University School of Medicine, USA) for providing the Notch RNAi line, K. Beebe for helpful advice and comments on the Drosophila intestinal studies, G. Lam for establishing the Drosophila MPC overexpression strain, O. Yilmaz and D. Sabatini for assistance and insight into intestinal stem cell metabolism, D. Tantin for critiques and comments, members of the Rutter laboratory for assistance and advice, J. O’Shea, R. Orbus and C. DeHeer for assistance with NanoString, W. Swiatek for mouse assistance, ARUP Institute for Clinical and Experimental Pathology, and S. R. Tripp and E. Hammond for histology; L. Nikolova at the University of Utah Electron Microscopy Core Laboratory performed electron microscopy; mass spectrometry analysis was performed at the Mass Spectrometry and Proteomics Core Facility at the University of Utah. Mass spectrometry equipment was obtained through NCRR Shared Instrumentation Grant no. 1 S10 RR020883-01, 1 S10 RR025532-01A1, NIH 1 S10OD021505-01 (J.E.C.) and the Diabetes and Metabolism Center at the University of Utah. This study was conducted with support from the Biorepository and Molecular Pathology Shared Resource supported by the Cancer Center Support Grant awarded to the Huntsman Cancer Institute by the National Cancer Institute of the National Institutes of Health. Nanostring transcript analysis utilized the Molecular Diagnostics Section of the Biorepository and Molecular Pathology Shared Resource and was supported by the National Cancer Institute of the National Institutes of Health under Award Number P30CA042014 (the content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH). J. Marvin at the University of Utah Flow Cytometry Facility carried out flow sorting (National Cancer Institute through Award Number 5P30CA042014-24, National Center for Research Resources of the National Institutes of Health under Award Number 1S10RR026802-01). Funding was also provided by HHMI (J.R.), Treadwell (J.R.) and RO1GM094232 (to J.R. and C.S.T.). J.C.S. was supported by an NIH Developmental Biology Training Grant (5T32 HD07491) and a University of Utah Graduate Research Fellowship. D.R.W. was supported by a University of Utah Graduate Research Fellowship.
Author information
Authors and Affiliations
Contributions
Conceptualization, J.C.S., D.R.W., C.S.T. and J.R.; methodology, J.C.S., D.R.W., C.B., H.Z., P.W., J.T., A.F., J.M., L.K.S., C.S.E., K.A.O., D.D., P.K., M.P.B., D.Y.L., J.E.C., H.R.C., W.E.L., C.S.T. and J.R.; investigation, J.C.S., D.R.W., C.B., H.Z., P.W., J.T., A.F., J.M., L.K.S., C.S.E., R.M., D.D. and P.K.; formal analysis, J.C.S., D.R.W., C.B., P.W., T.C.W., R.M., L.J., R.J.D. and J.E.C.; writing—original draft, review and editing, J.C.S., D.R.W., C.S.T. and J.R.; funding acquisition, C.S.T. and J.R.; resources, D.Y.L., J.C., C.S.T. and J.R.; supervision, C.S.T. and J.R.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Additional images of flow gating and IHC in mouse and human.
(a) Gating startegy for sorting of Lgr5-EGFP positive stem cells for transcript abundance. Top showing final gating for collection of GFP-low, GFP-mid, and GFP-high cells for transcript abundance. Bottom showing full gating strategy exclusion of cell debris and purification of singlet cells. (b) IHC of VDAC1 and MPC1 throughout the mouse small intestine from proximal to distal showing staining of crypt base and differentiating transient amplifying cells. (scale bar = 20 μm). (c) Human Protein Atlas (http://www.proteinatlas.org) images of duodenum, small intestine, and colon stained for TFAM, NRF1, PDK1, and CK20 (scale bar = 20 μm (b), 100 μm (c)).
Supplementary Figure 2 Drosophila MPC regulates stem cell proliferation.
(a) esg-GAL4 used to target RNAi as indicated or MPC overexpression (o/e) under infected conditions (scale bar = 50 μm). pHH3 + cells was quantified per intestine. (Control n = 10 (no infection) Control (with infection) n = 10, MPC O/E n = 10, LDH RNAi n = 10 MPC RNAi n = 10 PDH RNAi n = 10) (b) Western blot analysis to detect dMPC1 protein in control animals, dMPC1 mutants, HSP70-GAL4 controls, and HSP70 > UAS-dMPC1-dMPC2 transgenic animals following a 30-minute heat treatment and four hour recovery. Data are mean ± s.e.m. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001. All p values were calculated using Student’s t-test. Unprocessed western blots are provided in Supplementary Figure 7.
Supplementary Figure 3 MPC1 allele and additional quantification of in vivo phenotype.
(a) Schematic of MPC1 allele used for inducible knockout. (b) Close-up images of villi tip from MPC1 knockout stained for H&E, alcian blue, MPC1, VDAC and CK20 (scale bar = 20 μm). (c) Body weight for control and MPC1 knockout animals 30 days post-treatment. (n = 5 male control and knockout, n = 6 female control, = 3 knockout) (d) Total crypt + villi height and crypt height from individual regions of the small intestine. (n = 9 control, = 10 knockout for crypt + villi, n = 10 control, = 9 knockout for crypt height from each region) (e) Quantification of Alcian Blue positive cells per area of the proximal small intestine. (n = 10 control and knockout) (f) Quantification fo BrdU positive cells per crypt of proximal small intestine. (n = 9 control, = 10 knockout) (g) Images of Olfm4 and Lgr5 in situ in control and MPC1 knockout. (scale bar = 50μm) (h) Additional quantification of in situ staining. (n = 9 control, n = 10 knockout) (i) Quantification of Lrig1 IHC staining in control and MPC1 knockout proximal small intestine. (n = 9 control, n = 10 knockout) Data are mean ± s.e.m. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001. All p values were calculated using Student’s t-test.
Supplementary Figure 4 Additional characterization of in vito Mpc1 Lgr5-KO.
(a) Flow cytometry profile for organoids from control and MPC1 knockout cultures showing GFP positivity. (b) Long-term cultures retain increased growth ability throughout extended passaging (over ∼2 months) (n = 15 control and knockout, scale bar = 200 μm) (c) Control and MPC1 knockout organoid western blots for MPC1 and MPC2 and pyruvate dehydrogenase (phospho 293 and total). (d) Transcript abundance of MPC1 knockout organoids showing stem and differentiation marks (n = 3). (e) Quantification of specific Acetyl Histone H3 marks in control and knockout organoids (paired control set to 1.0) (n = 8 organoids). (f) Steady state metabolite abundance for control and MPC1 knockout organoids (n = 6 control and knockout). Data are mean ± s.e.m. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001. All p values were calculated using Student’s t-test. Unprocessed western blots are provided in Supplementary Figure 7.
Supplementary Figure 5 Effects of UK-5099 in maintaining organoids.
(a) Organoid formation from different regions of small intestine and colon with treatment conditions (n = 6 per condition, p values by t-test with respect to wENR for each location). (b) Loss of passaging efficiency in crypts maintained in wENR compared to UK-5099. When identical cell numbers are passaged normally, wENR treated are more likely to fail to passage. Images are of multiple matrigel/organoid spots of identical size (5uL) on a 6 well plate tiled to make one image (organoids from n = 3 wells were counted and number per well used for quantification, scale bar = 2000 μm). (c) Formation of hyperintense lumens in crypt organoids grown in wENR compared to UK-5099 3 days after changing media to remove CHIR99021 and Valproic acid (n = 69 total organoids from 6 individual wells). (d) Dose curves for individual drugs in supporting organoid formation from crypts (n = 8 wENR, all other conditions n = 6, p value by t-test with respect to wENR). Data are mean ± s.e.m. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001. All p values were calculated using Student’s t-test.
Supplementary Figure 6 Effects of UK-5099 and MPC over expression.
(a) Flux profiling of U-C13 Glucose incorporation into Citrate for tMEF and C2C12 cell lines treated with UK-5099 or Valproic Acid over 4 hours (n = 3 per condition per timepoint). (b) Representative images of infected Lgr5-EGFP (green) organoids with either Empty Vector-mCherry (in red) or MPC1-iRFP (in blue) and MPC2 mCherry (in red). Traces below represent intensity of each color over the length of the line outlined by white dotted line in the image. (n = 4 organoids with at least 15 infected cells per organoid counted). (c) ENCODE track for Myc and associated transcription factors localized to the Mpc1 promoter. (d) Model where crypt homeostasis involves low pyruvate oxidation at the base which increases during differentiation. MPC1 loss leads to reduced pyruvate oxidation, increased glycolysis, increased proliferation, and an expansion of the stem cell compartment within the crypt. Data are mean ± s.e.m. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001. All p values were calculated using Student’s t-test.
Supplementary information
Supplementary Information
Supplementary Information (PDF 23221 kb)
Supplementary Information
Life Sciences Reporting Summary (PDF 77 kb)
Supplementary Table 1
Supplementary Information (XLS 26 kb)
Supplementary Table 2
Supplementary Information (XLS 22 kb)
Supplementary Table 3
Supplementary Information (XLS 21 kb)
Supplementary Table 4
Supplementary Information (XLS 22 kb)
Supplementary Table 5
Supplementary Information (XLSX 42 kb)
Supplementary Table 6
Supplementary Information (XLSX 20 kb)
Supplementary Table 7
Supplementary Information (XLSX 57 kb)
Rights and permissions
About this article
Cite this article
Schell, J., Wisidagama, D., Bensard, C. et al. Control of intestinal stem cell function and proliferation by mitochondrial pyruvate metabolism. Nat Cell Biol 19, 1027–1036 (2017). https://doi.org/10.1038/ncb3593
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb3593
This article is cited by
-
FOXO transcription factors as mediators of stress adaptation
Nature Reviews Molecular Cell Biology (2024)
-
Hypoxic preconditioning accelerates the healing of ischemic intestinal injury by activating HIF-1α/PPARα pathway-mediated fatty acid oxidation
Cell Death Discovery (2024)
-
Evaluation of EGFR and COX pathway inhibition in human colon organoids of serrated polyposis and other hereditary cancer syndromes
Familial Cancer (2024)
-
Cholesterol biosynthesis modulates differentiation in murine cranial neural crest cells
Scientific Reports (2023)
-
Lactate-dependent transcriptional regulation controls mammalian eye morphogenesis
Nature Communications (2023)