Abstract
Cellular forces are crucial for many biological processes but current methods to image them have limitations with respect to data analysis, resolution and throughput. Here, we present a robust approach to measure mechanical cell–substrate interactions in diverse biological systems by interferometrically detecting deformations of an elastic micro-cavity. Elastic resonator interference stress microscopy (ERISM) yields stress maps with exceptional precision and large dynamic range (2 nm displacement resolution over a >1 μm range, translating into 1 pN force sensitivity). This enables investigation of minute vertical stresses (<1 Pa) involved in podosome protrusion, protein-specific cell–substrate interaction and amoeboid migration through spatial confinement in real time. ERISM requires no zero-force reference and avoids phototoxic effects, which facilitates force monitoring over multiple days and at high frame rates and eliminates the need to detach cells after measurements. This allows observation of slow processes such as differentiation and further investigation of cells, for example, by immunostaining.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Geiger, B., Spatz, J. P. & Bershadsky, A. D. Environmental sensing through focal adhesions. Nat. Rev. Mol. Cell Biol. 10, 21–33 (2009).
Koser, D. E. et al. Mechanosensing is critical for axon growth in the developing brain. Nat. Neurosci. 19, 1592–1598 (2016).
Kshitiz et al. Control of stem cell fate and function by engineering physical microenvironments. Integr. Biol. 4, 1008–1018 (2012).
Lo, C. M., Wang, H. B., Dembo, M. & Wang, Y. L. Cell movement is guided by the rigidity of the substrate. Biophys. J. 79, 144–152 (2000).
Plotnikov, S. V., Pasapera, A. M., Sabass, B. & Waterman, C. M. Force fluctuations within focal adhesions mediate ECM-rigidity sensing to guide directed cell migration. Cell 151, 1513–1527 (2012).
Paszek, M. J. et al. Tensional homeostasis and the malignant phenotype. Cancer Cell 8, 241–254 (2005).
Jannat, R. A., Robbins, G. P., Ricart, B. G., Dembo, M. & Hammer, D. A. Neutrophil adhesion and chemotaxis depend on substrate mechanics. J. Phys. Condens. Matter 22, 194117 (2010).
Ricart, B. G., Yang, M. T., Hunter, C. A., Chen, C. S. & Hammer, D. A. Measuring traction forces of motile dendritic cells on micropost arrays. Biophys. J. 101, 2620–2628 (2011).
Betz, T., Koch, D., Lu, Y.-B., Franze, K. & Käs, J. A. Growth cones as soft and weak force generators. Proc. Natl Acad. Sci. USA 108, 13420–13425 (2011).
Tomasek, J. J., Gabbiani, G., Hinz, B., Chaponnier, C. & Brown, R. A. Myofibroblasts and mechano-regulation of connective tissue remodelling. Nat. Rev. Mol. Cell Biol. 3, 349–363 (2002).
Mertz, A. F. et al. Scaling of traction forces with the size of cohesive cell colonies. Phys. Rev. Lett. 108, 198101 (2012).
Lemmon, C. A., Chen, C. S. & Romer, L. H. Cell traction forces direct fibronectin matrix assembly. Biophys. J. 96, 729–738 (2009).
Brugués, A. et al. Forces driving epithelial wound healing. Nat. Phys. 10, 683–690 (2014).
Tambe, D. T. et al. Collective cell guidance by cooperative intercellular forces. Nat. Mater. 10, 469–475 (2011).
Case, L. B. & Waterman, C. M. Integration of actin dynamics and cell adhesion by a three-dimensional, mechanosensitive molecular clutch. Nat. Cell Biol. 17, 955–963 (2015).
Labernadie, A. et al. Protrusion force microscopy reveals oscillatory force generation and mechanosensing activity of human macrophage podosomes. Nat. Commun. 5, 5343 (2014).
Bergert, M. et al. Force transmission during adhesion-independent migration. Nat. Cell Biol. 17, 524–529 (2015).
Liu, Y. J. et al. Confinement and low adhesion induce fast amoeboid migration of slow mesenchymal cells. Cell 160, 659–672 (2015).
Toyjanova, J., Flores-Cortez, E., Reichner, J. S. & Franck, C. Matrix confinement plays a pivotal role in regulating neutrophil-generated tractions, speed, and integrin utilization. J. Biol. Chem. 290, 3752–3763 (2015).
Harris, A. K., Wild, P. & Stopak, D. Silicone rubber substrata: a new wrinkle in the study of cell locomotion. Science 208, 177–179 (1980).
Burton, K. & Taylor, D. L. Traction forces of cytokinesis measured with optically modified elastic substrata. Nature 385, 450–454 (1997).
Dembo, M. & Wang, Y. L. Stresses at the cell-to-substrate interface during locomotion of fibroblasts. Biophys. J. 76, 2307–2316 (1999).
Schwarz, U. S. et al. Calculation of forces at focal adhesions from elastic substrate data: the effect of localized force and the need for regularization. Biophys. J. 83, 1380–1394 (2002).
Sabass, B., Gardel, M. L., Waterman, C. M. & Schwarz, U. S. High resolution traction force microscopy based on experimental and computational advances. Biophys. J. 94, 207–220 (2008).
Hur, S. S., Zhao, Y., Li, Y. S., Botvinick, E. & Chien, S. Live cells exert 3-dimensional traction forces on their substrata. Cell. Mol. Bioeng. 2, 425–436 (2009).
Legant, W. R. et al. Multidimensional traction force microscopy reveals out-of-plane rotational moments about focal adhesions. Proc. Natl Acad. Sci. USA 110, 881–886 (2013).
Álvarez-González, B. Three-dimensional balance of cortical tension and axial contractility enables fast amoeboid migration. Biophys. J. 108, 821–832 (2015).
Colin-York, H. et al. Super-resolved traction force microscopy (STFM). Nano Lett. 16, 2633–2638 (2016).
Tan, J. L. et al. Cells lying on a bed of microneedles: an approach to isolate mechanical force. Proc. Natl Acad. Sci. USA 100, 1484–1489 (2003).
Wolfenson, H. et al. Tropomyosin controls sarcomere-like contractions for rigidity sensing and suppressing growth on soft matrices. Nat. Cell Biol. 18, 33–42 (2015).
Polacheck, W. J. & Chen, C. S. Measuring cell-generated forces: a guide to the available tools. Nat. Methods 13, 415–423 (2016).
Polio, S. R., Rothenberg, K. E., Stamenovic, D. & Smith, M. L. A micropatterning and image processing approach to simplify measurement of cellular traction forces. Acta Biomater. 8, 82–88 (2012).
Bergert, M. et al. Confocal reference free traction force microscopy. Nat. Commun. 7, 12814 (2016).
Balaban, N. Q. et al. Force and focal adhesion assembly: a close relationship studied using elastic micropatterned substrates. Nat. Cell Biol. 3, 466–472 (2001).
Dimitriadis, E. K., Horkay, F., Maresca, J., Kachar, B. & Chadwick, R. S. Determination of elastic moduli of thin layers of soft material using the atomic force microscope. Biophys. J. 82, 2798–2810 (2002).
Levental, I., Georges, P. C. & Janmey, P. A. Soft biological materials and their impact on cell function. Soft Matter 2, 1–9 (2007).
Calle, Y., Burns, S., Thrasher, A. J. & Jones, G. E. The leukocyte podosome. Eur. J. Cell Biol. 85, 151–157 (2006).
Charras, G. & Paluch, E. Blebs lead the way: how to migrate without lamellipodia. Nat. Rev. Mol. Cell Biol. 9, 730–736 (2008).
Bastounis, E. et al. Both contractile axial and lateral traction force dynamics drive amoeboid cell motility. J. Cell Biol. 204, 1045–1061 (2014).
Lämmermann, T. et al. Rapid leukocyte migration by integrin-independent flowing and squeezing. Nature 453, 51–55 (2008).
Malawista, S. E. & de Boisfleury Chevance, A. Random locomotion and chemotaxis of human blood polymorphonuclear leukocytes (PMN) in the presence of EDTA: PMN in close quarters require neither leukocyte integrins nor external divalent cations. Proc. Natl Acad. Sci. USA 94, 11577–11582 (1997).
Friedl, P. & Weigelin, B. Interstitial leukocyte migration and immune function. Nat. Immunol. 9, 960–969 (2008).
Kinashi, T. Intracellular signalling controlling integrin activation in lymphocytes. Nat. Rev. Immunol. 5, 546–559 (2005).
Chen, J., Li, H., SundarRaj, N. & Wang, J. H.-C. α-smooth muscle actin expression enhances cell traction force. Cell Motil. Cytoskeleton 64, 248–257 (2007).
Delanoe-Ayari, H., Rieu, J. P. & Sano, M. 4D traction force microscopy reveals asymmetric cortical forces in migrating Dictyostelium cells. Phys. Rev. Lett. 105, 248103 (2010).
Alamo, J. C. del et al. Three-dimensional quantification of cellular traction forces and mechanosensing of thin substrata by Fourier traction force microscopy. PLoS ONE 8, e69850 (2013).
Renkawitz, J. et al. Adaptive force transmission in amoeboid cell migration. Nat. Cell Biol. 11, 1438–1443 (2009).
Petrie, R. J., Gavara, N., Chadwick, R. S. & Yamada, K. M. Nonpolarized signaling reveals two distinct modes of 3D cell migration. J. Cell Biol. 197, 439–455 (2012).
Yoshida, K. & Soldati, T. Dissection of amoeboid movement into two mechanically distinct modes. J. Cell Sci. 119, 3833–3844 (2006).
Kronenberg, N. M., Liehm, P. & Gather, M. C. Fabrication of elastic micro-cavity chips for use in Elastic Resonator Interference Stress Microscopy. Nat. Protoc. Exch. http://dx.doi.org/10.1038/protex.2017.052 (2017).
Graudejus, O., Görrn, P. & Wagner, S. Controlling the morphology of gold films on poly(dimethylsiloxane). ACS Appl. Mater. Interfaces 2, 1927–1933 (2010).
Fritz, J. L. & Owen, M. J. Hydrophobic recovery of plasma-treated polydimethylsiloxane. J. Adhes. 54, 33–45 (1995).
Bowden, N., Brittain, S., Evans, A. G., Hutchinson, J. W. & Whitesides, G. M. Spontaneous formation of ordered structures in thin films of metals supported on an elastomeric polymer. Nature 393, 146–149 (1998).
Li, B., Cao, Y.-P., Feng, X.-Q. & Gaoc, H. Mechanics of morphological instabilities and surface wrinkling in soft materials: a review. Soft Matter 8, 5728–5745 (2012).
Yuffa, A. J. & Scales, J. A. Object-oriented electrodynamic S-matrix code with modern applications. J. Comput. Phys. 231, 4823–4835 (2012).
Dimitriadis, E. K., Horkay, F., Maresca, J., Kachar, B. & Chadwick, R. S. Determination of elastic moduli of thin layers of soft material using the atomic force microscope. Biophys. J. 82, 2798–2810 (2002).
Pritchard, R. H., Lava, P., Debruyne, D. & Terentjev, E. M. Precise determination of the Poisson ratio in soft materials with 2D digital image correlation. Soft Matter 9, 6037–6045 (2013).
Acknowledgements
The authors thank K. Venkatesan Iyer and P. A. Reynolds for fruitful discussion, R. Shahapure for TFM reference measurements, A. L. Sobiech for illustrations and the DictyoBase for provision of AX3 strain Dictyostelium discoideum. This project has received funding from the Human Frontiers Science Program (RGY0074/2013), the Scottish Funding Council (via SUPA), the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement no. 640012), the EPSRC DTP (EP/L505079/1), the RS MacDonald Charitable Trust and the MRC (G1100116 and G110312/1).
Author information
Authors and Affiliations
Contributions
N.M.K., P.L. and M.C.G. developed ERISM. N.M.K. fabricated and characterized the micro-cavity substrates and conducted measurements. P.L. developed the data analysis, stress map calculation and graphical data presentation. A.S. contributed to protein coating, staining and cell culture. J.A.K. designed and prepared the primary mouse fibroblast experiment. J.G.B. prepared and assisted in the T cell experiment. G.S. and K.F. performed rheometry and TFM, and contributed to AFM and general discussion. S.J.P. proposed and prepared the macrophage experiment. M.C.G. supervised the project. N.M.K. and M.C.G. wrote the manuscript with input from all authors.
Corresponding author
Ethics declarations
Competing interests
The University of St Andrews has filed a patent application related to the technique described here.
Integrated supplementary information
Supplementary Figure 1 Development and characterization of ERISM micro-cavities and data analysis.
(a–d) Oxygen plasma treatment of silicone elastomer surface. (a) Increase in water contact angle for different plasma treatment durations at plasma power of 80–90 W. (b) Reflectance of 15 nm-thick gold layers evaporated on glass (blue) and on untreated elastomer (black). (The oscillation for the black line is due to thin film interference.) (c) Reflectance at 650 nm (left) and apparent Young’s modulus (right, calculated for an indentation force of 2 nN) of surface-oxidized elastomer layers with 15 nm top gold mirror for different plasma durations t and plasma powers P (process pressure, 3 mtorr; gas flow, 20 sccm O2 and 5 sccm Ar). The apparent Young’s modulus was obtained by AFM indentation measurements with a 17 μm-diameter glass sphere. For comparison the reflectance and Young’s modulus, respectively, of a pristine non-oxidized elastomer sample without gold coating are shown at the origins of the t–P planes. (d) Plot of apparent Young’s modulus versus reflectance for all samples shown in (c). The region where low reflectance renders ERISM measurements unreliable is marked in red. (e) Wavelength-dependent intensity of the probe light used for the ERISM measurement when incident on the bottom glass side of the micro-cavity through a 4×, 10×, 20× and 40× objective, respectively. (f) Measurement of the Young’s modulus of the elastomer within the micro-cavity obtained by comparing AFM indentation forces to FEM predictions obtained from ERISM deformation profiles. See Methods for details.
Supplementary Figure 2 Flow cytometry of isolated human macrophages.
Adherent cells from isolated peripheral blood mononuclear cells were recovered and stained with anti-CD14-PE, shown shaded, compared to auto-fluorescence from cells without antibody staining (unshaded).
Supplementary Figure 3 Demonstration of the sensitivity of ERISM to horizontally applied forces.
(a) The needle of an AFM cantilever is slightly indented into the ERISM chip to make firm contact and the cantilever is then pulled to the side to apply a defined lateral force. This gives rise to an asymmetric ERISM signal that grows with increasing horizontal force. (b) Peak ERISM signal detected versus horizontal force applied by AFM. Red line, linear fit (R2 > 0.99). Dashed lines, displacement resolution of ERISM in the present configuration and resulting minimal horizontal force that can be detected.
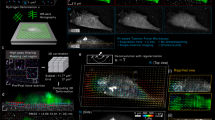
Supplementary Figure 4 High frame rate ERISM (a–b), ERISM for long-term time-lapse studies of the mechanical activity of cells during mesenchymal migration (c–e) and differentiation of fibroblasts into myofibroblasts in situ on the ERISM micro-cavity (f–g).
(a) Phase contrast image and ERISM map of 3T3 fibroblast cells. Images from an 11.5 min-ERISM time-lapse measurement with a frame acquired every 2.3 s. (b) Evolution of mean displacement under the cell nucleus and the lamellum, respectively (areas circled in blue and red in (a)). The black trace shows the evolution for an area where no cell is present (black circle in (a)). Also see Supplementary Video 5. (c) Phase contrast image and ERISM map for primary mouse fibroblasts after cells migrated out of a tissue explant directly placed on the micro-cavity substrate. (d) ERISM time-lapse series of fibroblast migration for the cell highlighted in (c). The cell boundary is indicated by a black line. (e) Temporal evolution of (left) the force applied in each direction with respect to the cell centre as defined in the first panel of (d) and (right) the total force and speed of cell migration. Position and size of white circles indicate direction and speed of migration. [A similar analysis of the moment of the force orientation forces has been performed for Dictyostelium discoideum using TFM, see H. Tanimoto and M. Sano, Biophys. J. 106, 16–25 (2014).] (f) Representative phase contrast image (left) and ERISM stress map (right) of a cell after 66 h of TGF-β treatment. Cell outline is shown in black. (g) Fluorescence images of immune-labelled α-SMA (green) after fixation and permeabilisation of cells on the micro-cavity. Left: Cells stimulated with TGF-β for 66 h. Right: Non-stimulated control cells kept on the micro-cavity for the same time. While cells in the control group show weak and unspecific background fluorescence, parallel arranged α-SMA fibres arching through the cell bodies are clearly visible in differentiated cells. Scale bars, (a) 50 μm, (c) 100 μm, (d,f) 50 μm and (g) 200 μm.
Supplementary information
Supplementary Information
Supplementary Information (PDF 753 kb)
Illustration of mechanical stability of the micro-cavities used for ERISM.
Real time video of monochromatic reflectance of a micro-cavity, into which a 17 μm dimeter glass bead (glued to the tip of an AFM probe) was indented by 900 nm. While keeping the indentation constant, the bead is moved horizontally across the micro-cavity. (AVI 5296 kb)
Fourier-filtered ERISM time-lapse investigation of podosome protrusions of a human macrophage.
Phase contrast microscopy (left) and spatial Fourier-filtered stress map (right). (AVI 7940 kb)
Fourier-filtered ERISM time-lapse investigation of podosome protrusion of a human macrophage during cell migration.
Phase contrast microscopy (top) and spatial Fourier filtered substrate displacement (bottom). (AVI 4644 kb)
ERISM time-lapse investigation of amoeboid migration in spatial confinement.
Video shows the substrate displacement caused by a Dictyostelium discoideum amoeba migrating through a 5 μm-thick void formed above the micro-cavity substrate. The outline of the amoeba is shown in black. (AVI 1899 kb)
High-speed ERISM time-lapse investigation of 3T3 fibroblasts.
Phase contrast microscopy (left) and substrate displacement in nm (right). The inset shows the displacement for the region highlighted by a white box in the bright-field and displacement images with increased contrast to show displacements due to expansion of the cell edge in more detail. A frame was taken every 2.3 s. (AVI 2699 kb)
Long-term ERISM time-lapse investigation of 3T3 fibroblasts.
Phase-contrast microscopy (left) and substrate displacement in nm (right). The video was recorded over 5.5 days and contains more than 1,600 individual ERISM maps. (AVI 15341 kb)
ERISM time-lapse investigation of mesenchymal migration in a culture of primary mouse fibroblasts.
Video shows an overlay of substrate displacement with corresponding phase contrast microscopy image. Pulling is indicated in red, pushing is indicated in blue. (AVI 7125 kb)
Close-up ERISM time-lapse of mesenchymal migration of individual primary mouse fibroblast.
Phase contrast microscopy (left) and substrate displacement (right). (AVI 642 kb)
Rights and permissions
About this article
Cite this article
Kronenberg, N., Liehm, P., Steude, A. et al. Long-term imaging of cellular forces with high precision by elastic resonator interference stress microscopy. Nat Cell Biol 19, 864–872 (2017). https://doi.org/10.1038/ncb3561
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb3561
This article is cited by
-
High-resolution assessment of multidimensional cellular mechanics using label-free refractive-index traction force microscopy
Communications Biology (2024)
-
Mechanisms and roles of podosomes and invadopodia
Nature Reviews Molecular Cell Biology (2023)
-
Imaging cellular forces with photonic crystals
Nature Communications (2023)
-
KIAA0319 influences cilia length, cell migration and mechanical cell–substrate interaction
Scientific Reports (2022)
-
Quantifying force transmission through fibroblasts: changes of traction forces under external shearing
European Biophysics Journal (2022)