Abstract
Growing evidence in vertebrates predicts that cellular haem levels in animals are maintained not only by a cell’s internal capacity for haem synthesis in a cell-autonomous manner, but also by an inter-organ haem trafficking network through cell-non-autonomous regulation. Using Caenorhabditis elegans, a genetically and optically amenable animal model for visualizing haem-dependent signalling, we show that HRG-7, a protein with homology to aspartic proteases, mediates inter-organ signalling between the intestine and extra-intestinal tissues. Intestinal HRG-7 functions as a secreted signalling factor during haem starvation in extra-intestinal tissues and is regulated through a DBL-1, homologous to BMP5, dependent signal from neurons. Given the evidence that vertebrate homologues exist for each of the components of the HRG-7-mediated signalling pathway, it is conceivable that the cell-non-autonomous signalling framework that we uncovered in C. elegans may have functional relevance for inter-organ regulation of iron and haem metabolism in humans.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
References
Hamza, I. Intracellular trafficking of porphyrins. ACS Chem. Biol. 1, 627–629 (2006).
Severance, S. & Hamza, I. Trafficking of heme and porphyrins in metazoa. Chem. Rev. 109, 4596–4616 (2009).
Korolnek, T. & Hamza, I. Macrophages and iron trafficking at the birth and death of red cells. Blood 125, 2893–2897 (2015).
Keel, S. B. et al. A heme export protein is required for red blood cell differentiation and iron homeostasis. Science 319, 825–828 (2008).
Cao, C. & O’Brien, K. O. Pregnancy and iron homeostasis: an update. Nutr. Rev. 71, 35–51 (2013).
Yang, Z. et al. Kinetics and specificity of feline leukemia virus subgroup C receptor (FLVCR) export function and its dependence on hemopexin. J. Biol. Chem. 285, 28874–28882 (2010).
Haldar, M. et al. Heme-mediated SPI-C induction promotes monocyte differentiation into iron-recycling macrophages. Cell 156, 1223–1234 (2014).
Yuan, X. et al. Regulation of intracellular heme trafficking revealed by subcellular reporters. Proc. Natl Acad. Sci. USA 113, E5144–E5152 (2016).
Sinclair, J. & Hamza, I. Lessons from bloodless worms: heme homeostasis in C. elegans. Biometals 28, 481–489 (2015).
Rao, A. U., Carta, L. K., Lesuisse, E. & Hamza, I. Lack of heme synthesis in a free-living eukaryote. Proc. Natl Acad. Sci. USA 102, 4270–4275 (2005).
Durieux, J., Wolff, S. & Dillin, A. The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell 144, 79–91 (2011).
Korolnek, T., Zhang, J., Beardsley, S., Scheffer, G. L. & Hamza, I. Control of metazoan heme homeostasis by a conserved multidrug resistance protein. Cell Metab. 19, 1008–1019 (2014).
Rajagopal, A. et al. Haem homeostasis is regulated by the conserved and concerted functions of HRG-1 proteins. Nature 453, 1127–1131 (2008).
Severance, S. et al. Genome-wide analysis reveals novel genes essential for heme homeostasis in Caenorhabditis elegans. PLoS Genet. 6, e1001044 (2010).
Nemeth, E. et al. Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science 306, 2090–2093 (2004).
Kim, B. E. et al. Cardiac copper deficiency activates a systemic signaling mechanism that communicates with the copper acquisition and storage organs. Cell Metab. 11, 353–363 (2010).
Kautz, L. et al. Identification of erythroferrone as an erythroid regulator of iron metabolism. Nat. Genet. 46, 678–684 (2014).
Sinclair, J. & Hamza, I. A novel heme response element mediates transcriptional regulation in Caenorhabditis elegans. J. Biol. Chem. 285, 39536–39543 (2010).
McGhee, J. D. et al. The ELT-2 GATA-factor and the global regulation of transcription in the C. elegans intestine. Dev. Biol. 302, 627–645 (2007).
Mahoney, T. R. et al. Intestinal signaling to GABAergic neurons regulates a rhythmic behavior in Caenorhabditis elegans. Proc. Natl Acad. Sci. USA 105, 16350–16355 (2008).
Oka, T., Toyomura, T., Honjo, K., Wada, Y. & Futai, M. Four subunit a isoforms of Caenorhabditis elegans vacuolar H+-ATPase. Cell-specific expression during development. J. Biol. Chem. 276, 33079–33085 (2001).
Pujol, N., Bonnerot, C., Ewbank, J. J., Kohara, Y. & Thierry-Mieg, D. The Caenorhabditis elegans unc-32 gene encodes alternative forms of a vacuolar ATPase a subunit. J. Biol. Chem. 276, 11913–11921 (2001).
Chen, C., Samuel, T. K., Sinclair, J., Dailey, H. A. & Hamza, I. An intercellular heme-trafficking protein delivers maternal heme to the embryo during development in C. elegans. Cell 145, 720–731 (2011).
Hedgecock, E. M., Culotti, J. G., Thomson, J. N. & Perkins, L. A. Axonal guidance mutants of Caenorhabditis elegans identified by filling sensory neurons with fluorescein dyes. Dev. Biol. 111, 158–170 (1985).
Mak, H. Y., Nelson, L. S., Basson, M., Johnson, C. D. & Ruvkun, G. Polygenic control of Caenorhabditis elegans fat storage. Nat. Genet. 38, 363–368 (2006).
Bishop, N. A. & Guarente, L. Two neurons mediate diet-restriction-induced longevity in C. elegans. Nature 447, 545–549 (2007).
Evans, D. & Blumenthal, T. Trans splicing of polycistronic Caenorhabditis elegans pre-mRNAs: analysis of the SL2 RNA. Mol. Cell. Biol. 20, 6659–6667 (2000).
Ding, L. & Candido, E. P. Association of several small heat-shock proteins with reproductive tissues in the nematode Caenorhabditis elegans. Biochem. J. 351, 13–17 (2000).
Balklava, Z., Pant, S., Fares, H. & Grant, B. D. Genome-wide analysis identifies a general requirement for polarity proteins in endocytic traffic. Nat. Cell Biol. 9, 1066–1073 (2007).
Poea-Guyon, S. et al. The V-ATPase membrane domain is a sensor of granular pH that controls the exocytotic machinery. J. Cell Biol. 203, 283–298 (2013).
Wang, H. et al. Neuropeptide secreted from a pacemaker activates neurons to control a rhythmic behavior. Curr. Biol. 23, 746–754 (2013).
Hung, W. L., Wang, Y., Chitturi, J. & Zhen, M. A Caenorhabditis elegans developmental decision requires insulin signaling-mediated neuron-intestine communication. Development 141, 1767–1779 (2014).
Sieburth, D. et al. Systematic analysis of genes required for synapse structure and function. Nature 436, 510–517 (2005).
Liang, J., Yu, L., Yin, J. & Savage-Dunn, C. Transcriptional repressor and activator activities of SMA-9 contribute differentially to BMP-related signaling outputs. Dev. Biol. 305, 714–725 (2007).
Savage-Dunn, C., Yu, L., Gill, K., Awan, M. & Fernando, T. Non-stringent tissue-source requirements for BMP ligand expression in regulation of body size in Caenorhabditis elegans. Genet. Res. (Camb) 93, 427–432 (2011).
Suzuki, Y. et al. A BMP homolog acts as a dose-dependent regulator of body size and male tail patterning in Caenorhabditis elegans. Development 126, 241–250 (1999).
Morita, K., Chow, K. L. & Ueno, N. Regulation of body length and male tail ray pattern formation of Caenorhabditis elegans by a member of TGF-β family. Development 126, 1337–1347 (1999).
Schultz, R. D., Bennett, E. E., Ellis, E. A. & Gumienny, T. L. Regulation of extracellular matrix organization by BMP signaling in Caenorhabditis elegans. PLoS ONE 9, e101929 (2014).
Liang, J. et al. The Caenorhabditis elegans schnurri homolog sma-9 mediates stage- and cell type-specific responses to DBL-1 BMP-related signaling. Development 130, 6453–6464 (2003).
Popovici, C. et al. Direct and heterologous approaches to identify the LET-756/FGF interactome. BMC Genomics 7, 105 (2006).
Persson, A. et al. Natural variation in a neural globin tunes oxygen sensing in wild Caenorhabditis elegans. Nature 458, 1030–1033 (2009).
Gray, J. M. et al. Oxygen sensation and social feeding mediated by a C. elegans guanylate cyclase homologue. Nature 430, 317–322 (2004).
Williamson, A. L. et al. A multi-enzyme cascade of hemoglobin proteolysis in the intestine of blood-feeding hookworms. J. Biol. Chem. 279, 35950–35957 (2004).
Brinkworth, R. I., Prociv, P., Loukas, A. & Brindley, P. J. Hemoglobin-degrading, aspartic proteases of blood-feeding parasites: substrate specificity revealed by homology models. J. Biol. Chem. 276, 38844–38851 (2001).
Turk, B., Turk, D. & Turk, V. Protease signalling: the cutting edge. EMBO J. 31, 1630–1643 (2012).
Rawlings, N. D., Waller, M., Barrett, A. J. & Bateman, A. MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 42, D503–D509 (2014).
Cousin, C., Bracquart, D., Contrepas, A. & Nguyen, G. Potential role of the (pro)renin receptor in cardiovascular and kidney diseases. J. Nephrol. 23, 508–513 (2010).
Glondu, M. et al. A mutated cathepsin-D devoid of its catalytic activity stimulates the growth of cancer cells. Oncogene 20, 6920–6929 (2001).
Ueno, E., Sakai, H., Kato, Y. & Yamamoto, K. Activation mechanism of erythrocyte cathepsin E. Evidence for the occurrence of the membrane-associated active enzyme. J. Biochem. 105, 878–882 (1989).
Li, J. M., Umanoff, H., Proenca, R., Russell, C. S. & Cosloy, S. D. Cloning of the Escherichia coli K-12 hemB gene. J. Bacteriol. 170, 1021–1025 (1988).
Redemann, S. et al. Codon adaptation-based control of protein expression in C. elegans. Nat. Methods 8, 250–252 (2011).
Finney, M. & Ruvkun, G. The unc-86 gene product couples cell lineage and cell identity in C. elegans. Cell 63, 895–905 (1990).
Acknowledgements
We thank A. Chisholm, K. Liu, T. Gumienny, A. Jose and D. Hall for critical discussions and advice; T. Fukushige for preparing the RNA-seq libraries and extensive discussions; National Bioresource Project and S. Mitani (Tokyo Women’s Medical University, Japan) for the hrg-7 strain; H. Fares (University of Arizona, USA) for strain NP97 (Punc−122::GFP). This work was supported by funding from the National Institutes of Health DK074797 and a supplement to DK074797 (I.H.); support was also provided by the Intramural Program of the National Institute of Diabetes and Digestive and Kidney Diseases (M.K.). The genome-wide RNAi screen was funded by the Roche Foundation for Anemia Research (I.H.).
Author information
Authors and Affiliations
Contributions
Experimental design and execution were as follows: C. elegans experiments J.S., K.P., T.S., S.B., X.Y., J.Z., K.M., M.K. and I.H.; homology modelling and RNA-seq analyses S.Y.; J.S. and I.H. wrote the manuscript. All authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
I.H. is the President and Founder of Rakta Therapeutics Inc. (College Park, MD), a company involved in the development of haem transporter-related diagnostics.
Integrated supplementary information
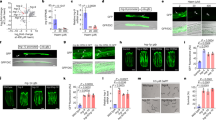
Supplementary Figure 1 Intestinal hrg-7 is a regulator of heme homeostasis.
(a) Top: ClustalW alignment of the hrg-7 5’ flanking region with homologous sequences in C. briggsae and C. remanei. White text with black background indicates a conserved nucleotide in all three species. Red box indicates HERE. Bottom: Alignment of the hrg-1 transcriptional regulatory region with the putative hrg-7 transcriptional regulatory region. Red box indicates HERE. White text with black background indicates conserved nucleotides in the HERE. Blue boxes indicate GATA sites. (b) GFP expression in strain IQ7701 (Phrg−7::GFP) grown in 4 or 20 μM heme. Scale bar = 20 μm. Images are representative of three independent experiments. (c) GFP expression in IQ7701 and IQ7702 (Phrg−7mut::GFP) in 4 μM heme. GFP was only detectable in IQ7701. Images are representative of three independent experiments. (d) Schematic showing that endogenous hrg-7 is knocked down by both hrg-7 dsRNA300bp and hrg-7 dsRNAORF, whereas the transgene hrg-7PR is resistant to knockdown by hrg-7 dsRNA300bp but not to knockdown by hrg-7 dsRNAORF. (e) Immunoblot analysis of HRG-7 and HRG-7PR from IQ6011 and IQ7711 fed dsRNA against control vector, hrg-7300bp, and hrg-7ORF. Membranes were probed with polyclonal anti-HRG-7 antibody and then incubated with HRP-conjugated anti-rabbit secondary antibody. ∗ indicates pro-HRG-7. + indicates mature HRG-7. Unprocessed blots are shown in Figure S6. Data is representative of two independent experiments.
Supplementary Figure 2 Secreted HRG-7 exhibits a tissue specific localization pattern.
(a) ZnMP staining in worms fed dsRNA against vector control, hrg-4, hrg-1, or hrg-7. Worms were exposed to RNAi from L1 to L4 larval stages, then pulsed with 10 mM ZnMP overnight. ZnMP staining was quantified using the region of interest function of Simple PCI. ZnMP intensity is presented as fold change compared to vector. Graph represents the mean and SEM of three biological independent experiments. N = 10 worms per treatment per experiment. ∗∗∗P < 0.001, ∗∗P < 0.01, ∗P < 0.05, when compared to control worms (one-way ANOVA). See Supplementary Table 3 for statistics source data. (b) mCherry expression in the anterior (left panel) and posterior (right panel) of strain IQ7670 (Pvha−7::HRG-7::mCherry). Scale bar = 20 μm. Images are representative of three independent experiments. (c) Coexpression of the coelomocyte marker Punc-122::GFP and Pvha−6::HRG-7::mCherry. Image depicts a middle coelomocyte. Scale bar = 20 μm. Images are representative of three independent experiments. (d) Left panels: mCherry expression in the gonad and coelomocytes of IQ8333 (Pvha−6::HRG-3::mCherry). No mCherry is observed in the anterior of the worm. Right panels: mCherry expression in the intestine, coelomocytes, and anterior of strain IQ8330 (Pvha−6::HRG-3-SP::mCherry). In the anterior of the worm, mCherry is non-specifically diffused throughout the pseudocoelomic fluid. Scale bar = 20 μm. Images are representative of three independent experiments. (e) Representative images of tissue specific expression of GFP/YFP markers Pceh−22::GFP (pharyngeal muscle), Pdpy−7::YFP (hypodermis), and Pmyo−2::GFP (body wall muscle) crossed into strain IQ7670 (Pvha−6::HRG-7::mCherry). ∗ indicates body wall muscle (BWM). Arrow indicates pharyngeal muscle (PM). Arrowhead indicates hypodermis (H). Scale bar = 20 μm. Images are representative of three independent experiments. (f) GFP fluorescence quantified from IQ6015 [(Phrg−1::GFP; lin-15b(n744)] fed dsRNA against control, hrg-4, mrp-5, or hrg-7 alone or in combination. GFP was quantified using COPAS BioSort. GFP is presented as fold change compared to vector. Graph represents the mean and SEM of three biological independent experiments. N = 120 worms per treatment per experiment. ∗∗∗P < 0.001, ∗∗P < 0.01, ∗P < 0.05 (one-way ANOVA). See Supplementary Table 3 for statistics source data.
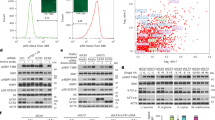
Supplementary Figure 3 Functional HRG-7 cannot be expressed by extraintestinal tissues.
(a) Images of strain IQ7170 (Phsp−16.2::HRG-7-mCherry) at timepoint = 0 and 360 minutes after heat shock. Coelomocytes are outlined by a yellow circle. Scale bar = 10 μm. (b) Fluorescence images of worms expressing hrg-7PR: :ICS::mCherry: from the vha-6 (intestine), myo-3 (muscle), or unc-119 (neuron) promoter. (c) Immunoblot analysis of HRG-7PR expressed from the vha-6, myo-3, or unc-119 promoter. Transgenic worms were fed dsRNA against hrg-7300bp. Membranes were probed with polyclonal anti-HRG-7 antibody and then incubated with HRP-conjugated anti-rabbit secondary antibody. * indicates pro-HRG-7. + indicates mature HRG-7. Unprocessed blots are shown in Figure S6. Data is representative of two independent experiments. (d) GFP fluorescence quantified from IQ6011 (Phrg−1::GFP) or worms expressing Phrg−1::GFP and HRG-7PR from the vha-6, myo-3, or unc-119 promoter fed dsRNA against control vector, hrg-7300bp, and hrg-7ORF at 10 μM heme. GFP was quantified using COPAS BioSort. GFP is presented as fold change compared to vector. Graph represents the mean and SEM of three biological independent experiments. N = 120 worms per treatment per experiment. ∗∗∗P < 0.001, ∗∗P < 0.01, ∗P < 0.05 (one-way ANOVA)). See Supplementary Table 3 for statistics source data.
Supplementary Figure 4 Transgenic HRG-7 rescues hrg-7 mutant phenotypes.
(a) Cartoon depicting the deleted region of hrg-7 in the tm6801 allele. Deletion is depicted by an underline. Open rectangles, exons; gray boxes, untranslated regions. (b) Homology model of HRG-7 using I-TASSER. Left: the putative active site is indicated by a red circle. The conserved disulfide bonds are indicated by white arrows. Alpha helixes are colored fuchsia. Beta sheets are colored yellow. Right: Zoomed in image of the putative active site. Yellow arrows indicate the catalytic aspartic acid residues and the conserved tyrosine of the flap region. (c) Immunoblot analysis of HRG-7 expression in worms expressing WT HRG-7 or HRG-7TM6801 and fed dsRNA against control vector or hrg-7. Membranes were probed with polyclonal anti-HRG-7 antibody and then incubated with HRP-conjugated anti-rabbit secondary antibody. * indicates pro-HRG-7. + indicates mature HRG-7. “Low” indicates low sensitivity detection. “High” indicates high sensitivity detection. Unprocessed blots are shown in Figure S6. Data is representative of three independent experiments. (d) GFP fluorescence quantified from IQ6011 expressing WT hrg-7, hrg-7(tm6801), or hrg-7(tm6801) and transgenic Pvha−6::hrg-7PR grown on OP50. GFP is presented as fold change compared to wild type (WT) worms. ∗∗∗P < 0.001, ∗∗P < 0.01, ∗P < 0.05 (one-way ANOVA). Graph represents the mean and SEM of three biological independent experiments. N = 120 worms per treatment per experiment. See Supplementary Table 3 for statistics source data. (e) Immunoblot analysis of titrated endogenous HRG-7 and HRG-7PR(D90A/D318A) expressed in the hrg-7(tm6801) background. Membranes were probed with polyclonal anti-HRG-7 antibody and then incubated with HRP-conjugated anti-rabbit secondary antibody. Irrelevant lanes were removed from the image. Unprocessed blots are shown in Figure S6. Images are representative of two independent experiments.
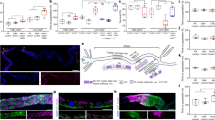
Supplementary Figure 5 Extra-intestinal regulation of intestinal hrg-1.
(a) GFP expression in strain BW1946 (Pdbl−1::GFP). Arrowhead indicates head neurons. Arrow indicates tail neurons. ∗ indicates body neuron. Scale bar = 100 μm (top panel); Scale bar = 20 μm (middle and lower panels). Images are representative of three independent experiments. (b) GFP quantification from IQ6011, IQ6311, and IQ6312 grown on OP50 supplemented with indicated heme concentrations. GFP fluorescence was quantified using COPAS BioSort. GFP is presented as relative expression on a scale from 1-100. Graph represents the mean and SEM of 50 worms per strain per heme concentration combined from three biological independent experiments. ∗∗∗P < 0.001, ∗∗P < 0.01, ∗P < 0.05 compared to WT worms (two-way ANOVA). (c) GFP-DBL-1 expression in strain VC1478. GFP signal was detected in neurons and neuron support cells. “hn” indicates head neuron. “sc” indicates support cells. “cn” indicates ventral nerve cord neuron. Scale bar = 20 μm. Images are representative of three independent experiments. (d) wCherry expression in strain RW10745 (Psma−9::HIS-24-wCherry). Arrow indicates hypodermal nuclei. Arrowhead and asterisks indicate intestinal and body neuron nuclei, respectively. Scale bar = 100 μm (top panel); Scale bar = 20 μm (middle and lower panels). Images are representative of three independent experiments. (e) GFP fluorescence quantified from IQ6011 expressing WT hrg-7 or hrg-7(tm6801) and fed dsRNA against vector, let-756, or snap-29. GFP was quantified using COPAS BioSort. GFP is presented as fold change compared to vector. Graph represents the mean and SEM of 1000 worms per treatment combined from four biological independent experiments. ∗∗∗P < 0.001, ∗∗P < 0.01, ∗P < 0.05 (Dunnett’s test).
Supplementary Figure 6 Unprocessed western blot scans.
Unprocessed western blots from Figures 1c, 3d, 3e, and Supplementary Figures 1e, 3c, 4c, 4e.
Supplementary information
Supplementary Information
Supplementary Information (PDF 5994 kb)
Supplementary Table 1
Supplementary Information (XLSX 10 kb)
Supplementary Table 2
Supplementary Information (XLSX 17 kb)
Supplementary Table 3
Supplementary Information (XLSX 23 kb)
Supplementary Table 4
Supplementary Information (XLSX 18 kb)
Rights and permissions
About this article
Cite this article
Sinclair, J., Pinter, K., Samuel, T. et al. Inter-organ signalling by HRG-7 promotes systemic haem homeostasis. Nat Cell Biol 19, 799–807 (2017). https://doi.org/10.1038/ncb3539
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb3539
This article is cited by
-
HRG-9 homologues regulate haem trafficking from haem-enriched compartments
Nature (2022)
-
Hc-hrg-2, a glutathione transferase gene, regulates heme homeostasis in the blood-feeding parasitic nematode Haemonchus contortus
Parasites & Vectors (2020)
-
Machinery for fungal heme acquisition
Current Genetics (2020)
-
Bovine neutrophils form extracellular traps in response to the gastrointestinal parasite Ostertagia ostertagi
Scientific Reports (2018)
-
Inter-organ regulation of haem homeostasis
Nature Cell Biology (2017)