Abstract
Self-renewing naive mouse embryonic stem cells (mESCs) contain few mitochondria, which increase in number and volume at the onset of differentiation. KBP (encoded by Kif1bp) is an interactor of the mitochondrial-associated kinesin Kif1Bα. We found that TDH, responsible for mitochondrial production of acetyl-CoA in mESCs, and the acetyltransferase GCN5L1 cooperate to acetylate Lys501 in KBP, allowing its recognition by and degradation via Fbxo15, an F-box protein transcriptionally controlled by the pluripotency core factors and repressed following differentiation. Defects in KBP degradation in mESCs result in an unscheduled increase in mitochondrial biogenesis, enhanced respiration and ROS production, and inhibition of cell proliferation. Silencing of Kif1Bα reverts the aberrant increase in mitochondria induced by KBP stabilization. Notably, following differentiation, Kif1bp−/− mESCs display impaired expansion of the mitochondrial mass and form smaller embryoid bodies. Thus, KBP proteolysis limits the accumulation of mitochondria in mESCs to preserve their optimal fitness, whereas KBP accumulation promotes mitochondrial biogenesis in differentiating cells.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Accession codes
References
Dyall, S. D., Brown, M. T. & Johnson, P. J. Ancient invasions: from endosymbionts to organelles. Science 304, 253–257 (2004).
Giorgi, C. et al. Mitochondrial Ca2+ and apoptosis. Cell Calcium 52, 36–43 (2012).
Xu, X. et al. Mitochondrial regulation in pluripotent stem cells. Cell Metab. 18, 325–332 (2013).
Rafalski, V. A., Mancini, E. & Brunet, A. Energy metabolism and energy-sensing pathways in mammalian embryonic and adult stem cell fate. J. Cell Sci. 125, 5597–5608 (2012).
Rehman, J. Empowering self-renewal and differentiation: the role of mitochondria in stem cells. J. Mol. Med. 88, 981–986 (2010).
Wanet, A., Arnould, T., Najimi, M. & Renard, P. Connecting mitochondria, metabolism, and stem cell fate. Stem Cells Dev. 24, 1957–1971 (2015).
Lin, J. et al. Transcriptional co-activator PGC-1α drives the formation of slow-twitch muscle fibres. Nature 418, 797–801 (2002).
Spiegelman, B. M. Transcriptional control of energy homeostasis through the PGC1 coactivators. Novartis Found. Symp. 286, 3–6 (2007).
Tokuzawa, Y. et al. Fbx15 is a novel target of Oct3/4 but is dispensable for embryonic stem cell self-renewal and mouse development. Mol. Cell. Biol. 23, 2699–2708 (2003).
Takahashi, K. & Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 (2006).
Skaar, J. R., Pagan, J. K. & Pagano, M. Mechanisms and function of substrate recruitment by F-box proteins. Nat. Rev. Mol. Cell Biol. 14, 369–381 (2013).
Chen, B. B. et al. E3 ligase subunit Fbxo15 and PINK1 kinase regulate cardiolipin synthase 1 stability and mitochondrial function in pneumonia. Cell Rep. 7, 476–487 (2014).
Brooks, A. S. et al. Homozygous nonsense mutations in KIAA1279 are associated with malformations of the central and enteric nervous systems. Am. J. Hum. Genet. 77, 120–126 (2005).
Lyons, D. A., Naylor, S. G., Mercurio, S., Dominguez, C. & Talbot, W. S. KBP is essential for axonal structure, outgrowth and maintenance in zebrafish, providing insight into the cellular basis of Goldberg-Shprintzen syndrome. Development 135, 599–608 (2008).
Hirokawa, N., Niwa, S. & Tanaka, Y. Molecular motors in neurons: transport mechanisms and roles in brain function, development, and disease. Neuron 68, 610–638 (2010).
Wozniak, M. J., Melzer, M., Dorner, C., Haring, H. U. & Lammers, R. The novel protein KBP regulates mitochondria localization by interaction with a kinesin-like protein. BMC Cell Biol. 6, 35 (2005).
Kevenaar, J. T. et al. Kinesin-binding protein controls microtubule dynamics and cargo trafficking by regulating kinesin motor activity. Curr. Biol. 26, 849–861 (2016).
Drerup, C. M., Lusk, S. & Nechiporuk, A. Kif1B interacts with KBP to promote axon elongation by localizing a microtubule regulator to growth cones. J. Neurosci. 36, 7014–7026 (2016).
Still, A. J. et al. Quantification of mitochondrial acetylation dynamics highlights prominent sites of metabolic regulation. J. Biol. Chem. 288, 26209–26219 (2013).
Scott, I., Webster, B. R., Li, J. H. & Sack, M. N. Identification of a molecular component of the mitochondrial acetyltransferase programme: a novel role for GCN5L1. Biochem J. 443, 655–661 (2012).
Webster, B. R. et al. Restricted mitochondrial protein acetylation initiates mitochondrial autophagy. J. Cell Sci. 126, 4843–4849 (2013).
Scott, I. et al. GCN5-like protein 1 (GCN5L1) controls mitochondrial content through coordinated regulation of mitochondrial biogenesis and mitophagy. J. Biol. Chem. 289, 2864–2872 (2014).
Wang, J. et al. Dependence of mouse embryonic stem cells on threonine catabolism. Science 325, 435–439 (2009).
Shyh-Chang, N. et al. Influence of threonine metabolism on S-adenosylmethionine and histone methylation. Science 339, 222–226 (2013).
Kim, J., Chu, J., Shen, X., Wang, J. & Orkin, S. H. An extended transcriptional network for pluripotency of embryonic stem cells. Cell 132, 1049–1061 (2008).
Buecker, C. et al. Reorganization of enhancer patterns in transition from naive to primed pluripotency. Cell Stem Cell 14, 838–853 (2014).
Alexander, P. B., Wang, J. & McKnight, S. L. Targeted killing of a mammalian cell based upon its specialized metabolic state. Proc. Natl Acad. Sci. USA 108, 15828–15833 (2011).
Friedman, J. R., Webster, B. M., Mastronarde, D. N., Verhey, K. J. & Voeltz, G. K. ER sliding dynamics and ER-mitochondrial contacts occur on acetylated microtubules. J. Cell Biol. 190, 363–375 (2010).
Prowse, A. B. et al. Analysis of mitochondrial function and localisation during human embryonic stem cell differentiation in vitro. PLoS ONE 7, e52214 (2012).
Karbowski, M. et al. Opposite effects of microtubule-stabilizing and microtubule-destabilizing drugs on biogenesis of mitochondria in mammalian cells. J. Cell Sci. 114, 281–291 (2001).
Zhou, W. et al. HIF1α induced switch from bivalent to exclusively glycolytic metabolism during ESC-to-EpiSC/hESC transition. EMBO J. 31, 2103–2116 (2012).
Carbognin, E., Betto, R. M., Soriano, M. E., Smith, A. G. & Martello, G. Stat3 promotes mitochondrial transcription and oxidative respiration during maintenance and induction of naive pluripotency. EMBO J. 35, 618–634 (2016).
Pauklin, S., Pedersen, R. A. & Vallier, L. Mouse pluripotent stem cells at a glance. J. Cell Sci. 124, 3727–3732 (2011).
Narendra, V. et al. CTCF establishes discrete functional chromatin domains at the Hox clusters during differentiation. Science 347, 1017–1021 (2015).
Young, L. M. et al. TIMELESS forms a complex with PARP1 distinct from its complex with TIPIN and plays a role in the DNA damage response. Cell Rep. 13, 451–459 (2015).
Washburn, M. P., Wolters, D. & Yates, J. R. III Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 19, 242–247 (2001).
Florens, L. et al. Analyzing chromatin remodeling complexes using shotgun proteomics and normalized spectral abundance factors. Methods 40, 303–311 (2006).
MacCoss, M. J. et al. Shotgun identification of protein modifications from protein complexes and lens tissue. Proc. Natl Acad. Sci. USA 99, 7900–7905 (2002).
Eng, J. K., McCormack, A. L. & Yates, J. R. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5, 976–989 (1994).
Tabb, D. L., McDonald, W. H. & Yates, J. R. III DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J. Proteome Res. 1, 21–26 (2002).
Paoletti, A. C. et al. Quantitative proteomic analysis of distinct mammalian Mediator complexes using normalized spectral abundance factors. Proc. Natl Acad. Sci. USA 103, 18928–18933 (2006).
Zybailov, B. et al. Statistical analysis of membrane proteome expression changes in Saccharomyces cerevisiae. J. Proteome Res. 5, 2339–2347 (2006).
D’Angiolella, V. et al. Cyclin F-mediated degradation of ribonucleotide reductase M2 controls genome integrity and DNA repair. Cell 149, 1023–1034 (2012).
Kuchay, S. et al. FBXL2- and PTPL1-mediated degradation of p110-free p85β regulatory subunit controls the PI(3)K signalling cascade. Nat. Cell Biol. 15, 472–480 (2013).
Ran, F. A. et al. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 (2013).
Sommer, C. A. et al. Induced pluripotent stem cell generation using a single lentiviral stem cell cassette. Stem Cells 27, 543–549 (2009).
Pagan, J. K. et al. Degradation of Cep68 and PCNT cleavage mediate Cep215 removal from the PCM to allow centriole separation, disengagement and licensing. Nat. Cell Biol. 17, 31–43 (2015).
Giorgi, C. et al. p53 at the endoplasmic reticulum regulates apoptosis in a Ca2+-dependent manner. Proc. Natl Acad. Sci. USA 112, 1779–1784 (2015).
Dankert, J. F. et al. Cyclin F-mediated degradation of SLBP limits H2A.X accumulation and apoptosis upon genotoxic stress in G2. Mol. Cell 64, 507–519 (2016).
Parkhomchuk, D. et al. Transcriptome analysis by strand-specific sequencing of complementary DNA. Nucleic Acids Res. 37, e123 (2009).
Acknowledgements
The authors thank C. Haynes, J. K. Pagan and T. Pozzan for critical reading of the manuscript; I. Aifantis (NYU School of Medicine, USA), R. Lammers (University of Tubingen, Germany), M. N. Sack (National Institutes of Health, USA) and F. Zhang (Massachusetts Institute of Technology, USA) for reagents. M.P. is grateful to T. M. Thor and T. B. Balduur for continuous support. V.D. is grateful to F. Catalanotti, G. Donato and A. Donato for continuous support. P.P. is grateful to C. d. Scrovegni for providing continuous support. This work was funded by grants from the National Institutes of Health (R37-CA076584, R01-GM057587 and R21-CA161108) and New York State Health Department (NYSTEM-N11G-255) to M.P.; the American-Italian Cancer Foundation and the National Institutes of Health (5T32CA009161-40) to V.D.; and The Italian Ministry of Health and the Italian Ministry of Education, University and Research (COFIN no. 20129JLHSY_002, FIRB no. RBAP11FXBC_002, and Futuro in Ricerca no. RBFR10EGVP_001), Telethon (GGP15219/B), the Italian Cystic Fibrosis Research Foundation (19/2014) and the Italian Association for Cancer Research (IG-18624) and local funds from the University of Ferrara and the Italian Ministry of Health to P.P. The authors also thank the NYU Genome Technology Center (partially funded by the Perlmutter Cancer Center Support Grant P30CA016087) for expert library preparation and sequencing for RNA-seq, and the NYU Applied Bioinformatics Laboratories for providing bioinformatics support and helping with the analysis and interpretation of the RNA-seq data using computing resources at the NYU High Performance Computing Facility. A.S., L.F. and M.P.W. are supported by the Stowers Institute for Medical Research. M.P. is an Investigator with the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Contributions
V.D. conceived the project, planned and performed most experiments, and co-wrote the manuscript. M.P. directed and coordinated the study, oversaw the results, and co-wrote the manuscript. M.B. and P.P. performed analyses of mitochondrial morphology and physiology. D.Simoneschi generated the CRISPR knockout mESC clones and helped with biochemical experiments and with RNA-seq data analysis. D.Sartini helped with some biochemical experiments. Y.K. performed the Fbxo15+/+ and Fbxo15−/− mouse breeding. M.S. provided reagents and advice. A.S., L.F. and M.P.W. performed the mass spectrometry analyses of the purifications performed by V.D. All authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Multidimensional Protein Identification Technology analysis of the Fbxo15 complex and its validation, and generation of Fbxo15−/− mESCs by CRISPR Genome Editing.
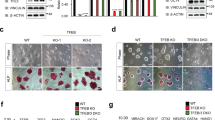
(a) HEK293T cells were transfected with either Strep-FLAG-tagged human Fbxo15 or Strep-FLAG-tagged human Fbxo21 (the latter used as control). The table shows the mass spectrometry analysis of the two immunopurifications, listing normalized spectral abundance factors (NSAFs) for the indicated proteins. The complete list of Fbxo15 interactors is reported in Supplementary Table 1. This experiment was performed twice. (b) HEK293T cells were transfected with an empty vector (EV) or constructs for the indicated human proteins. Whole cell extracts (WCE) were immunoprecipitated (IP) with anti-FLAG resin, and immunocomplexes were immunoblotted as indicated. (c) mESCs were transfected with either an siRNA to Fbxo15 (oligo #1) or a nontargeting (N/T) siRNA. Protein extracts were then immunoblotted for the indicated proteins. (d) mESCs were transfected with either a nontargeting (N/T) siRNA or siRNA to Fbxo15 and treated for 4 h with MG132, MLN4924, or DMSO. Cells were then collected and lysed for immunoblotting as indicated. The Nedd8-activating enzyme inhibitor MLN4924 blocks the neddylation of Cul1, which is required for the SCF activity. p27, an established substrate of the proteasome and the F-box protein Skp2, is used as positive control for MG132 and MLN4924, and as a negative control for Fbxo15 silencing. (e) mESCs were transfected with either an siRNA to Fbxo15 (oligo #1) or a nontargeting (N/T) siRNA. Cells were treated with cycloheximide (CHX) for the indicated times and protein extracts were then immunoblotted for the indicated proteins. Long (l. e.) and short (s. e.) exposures are shown for KBP. (f) Schematic representation of Fbxo15 genomic locus and gRNA target location. Exon 1 refers to mouse Fbxo15 gene in NC_000084.6 Reference GRCm38.p4 C57BL/6J and in NM_015798.3. In the bottom panel, full-length wild-type genomic DNA template and truncated mutant sequences identified by TOPO-TA cloning of Fbxo15 PCR from two independent mESCs knockout clones are depicted. Clone #1 sequence and chromatogram revealed two different deletion events; one allele contains a 31bp deletion, while the other allele contains a 16bp deletion. Clone #2 sequence and chromatogram identified a 14bp deletion event at both alleles. Unprocessed original scans of blots are shown Supplementary Fig. 9. Unless otherwise noted, experiments were performed at least three times.
Supplementary Figure 2 Identification of the KBP degron.
(a–d) HEK293T cells were transfected with either an empty vector (EV), wild type human KBP, or the indicated FLAG-tagged human KBP mutants (either deletion, truncation, or point mutation mutants, as indicated). Whole cell extracts (WCE) were immunoprecipitated (IP) with an anti-FLAG resin, and immunoprecipitates were probed with the indicated antibodies. (e) Alignment of the Fbxo15 binding motif in KBP orthologs. The critical lysine required for Fbxo15 binding is highlighted in red. (f) Schematic representation of KBP mutants. KBP mutants found to interact with Fbxo15 are indicated by the symbol (+). (±) denotes reduced binding, and (−) denotes a lack of binding. (g) HEK293T cells were transfected with either an empty vector (EV), wild type human KBP, or the indicated FLAG-tagged human KBP point mutation mutants. Whole cell extracts (WCE) were immunoprecipitated (IP) with an anti-FLAG resin, and immunoprecipitates were probed with the indicated antibodies. (h) Strep-FLAG-tagged human KBP was immunoprecipitated from HEK293T cells and subjected to PTM analysis. The table lists the number of spectra matching acetylated and phosphorylated peptides as a fraction of the total spectra detected for peptides bearing each modified residue and their corresponding modification percentage. This experiment was performed once. (i) HEK293T cells were transfected with either an empty vector (EV) or vectors expressing FLAG-tagged wild type human KBP or FLAG-tagged human KBP(KK/RR). Whole cell extracts were immunoprecipitated (IP) with anti-FLAG resin, and proteins were immunoblotted as indicated. Unprocessed original scans of blots are shown Supplementary Fig. 9. Unless otherwise noted, experiments were performed at least three times.
Supplementary Figure 3 GCN5L1 and TDH allow KBP degradation via Fbxo15.
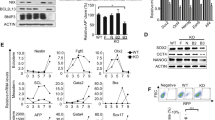
(a) mESCs were transfected with either a nontargeting (N/T) siRNA or siRNAs to Fbxo15 (oligo #2), GCN5L1, or KBP. Cells were either maintained in LIF-containing medium or induced to differentiate for 24 h after LIF withdrawal and exposure to retinoic acid (RA). Cells were then collected and lysed for immunoblotting as indicated. (b) mESCs were treated for 12 h with either DMSO or the TDH inhibitor Qc-1. Protein extracts were then immunoblotted for the indicated proteins. (c)The experiment was performed as in b, except that cells were treated with cycloheximide (CHX) for the indicated times. (d) mESCs were transfected with either a nontargeting (N/T) siRNA or siRNAs to Fbxo15 (oligo #1), GCN5L1, KBP, or TDH. Cells were either maintained in LIF-containing medium or induced to differentiate for 24 h after LIF withdrawal and exposure to neurobasal (NB) medium on non-adherent petri dishes. Cells were then collected and lysed for immunoblotting as indicated. Unprocessed original scans of blots are shown Supplementary Fig. 9. Unless otherwise noted, experiments were performed at least three times.
Supplementary Figure 4 Accumulation of KBP does not induce any major transcriptional rearrangement.
mESCs were infected with lentiviruses expressing either HA-tagged wild type mouse KBP or HA-tagged mouse KBP(KK/RR) and maintained in self renewing conditions. RNA-seq analysis was performed and results are shown as a MA plot of log2 fold changes versus the mean of normalized counts. Genes were considered differentially expressed when the P adjusted value was <0.05. ∼290 genes displayed differential expression, with modest fold changes (the relative expression for each affected transcript is comprised between −0.2 and +0.3). Neither a variation in the expression of the pluripotency core factors nor changes in the major metabolic drivers were detected. The complete list of FPKM reads is reported in Supplementary Table 2. n = 3 biologically independent experiments.
Supplementary Figure 5 Inhibition of KBP degradation promotes mitochondria-to-microtubule interaction and mitochondrial network expansion in mESCs.
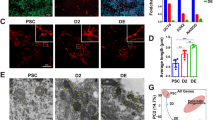
(a) mESCs were transfected with either a nontargeting (N/T) siRNA or an siRNA to Fbxo15 (oligo #1) and immunostained for TOM20 and β-tubulin. Microtubule-mitochondria co-localization was detected using confocal microscopy. Scale bar, 15 μm. In the bottom graph, the Manders’ Overlap Coefficient is shown. n = 61 and 40 cells (N/T and Fbxo15 siRNA, respectively). P value was calculated by unpaired t-test. Error bars indicate ± s.d. (b) mESCs were treated for 12 h with DMSO or Qc-1 and immunostained for TOM20 and β-tubulin. Microtubule-mitochondria co-localization was detected by confocal microscopy. Scale bar = 15 μm. In the bottom graph, the Manders’ Overlap Coefficient is shown. n = 32 and 47 cells (DMSO and Qc-1 treatment, respectively). P values were calculated by unpaired t-test. Error bars indicate ± s.d. (c) mESCs were either maintained in LIF-containing medium or induced to differentiate for 24 h after LIF withdrawal and exposure to retinoic acid (RA). Cells were stained with Mitotracker and DAPI and then analyzed by 3D deconvolution microscopy. Representative MIPs are displayed. Scale bar = 15 μm. (d–e) mESCs were transfected with either a nontargeting (N/T) siRNA or an siRNA to Fbxo15 (oligo #1) in the presence or absence of an siRNA to KBP, and maintained in LIF-containing medium. Cells were stained with Mitotracker and DAPI prior to fixation and then analyzed by confocal microscopy. Panel (d) shows the MIPs of representative cells (scale bar = 15 μm). Panel (e) shows the quantification of average mitochondrial content. n = 62, 62 and 33 cells (N/T, Fbxo15 and Fbxo15 + KBP siRNA, respectively). P value was calculated by one-way ANOVA. Error bars indicate ± s.d. (f) mESCs were transfected with either an siRNA to Fbxo15 (oligo #1) or a nontargeting (N/T) siRNA. Total cellular DNA was extracted and mtDNA copy number was evaluated by qPCR normalized using β-actin DNA copy number. mtDNA amount in mESCs expressing transfected with a nontargeting (N/T) siRNA was set as 100 and the graph shows the fold change. P value was calculated by t-test. Error bars indicate ± s.d. n = 4 biologically independent experiments (see Supplementary Table 6). (g) mESCs were transfected with either an siRNA to Fbxo15 (oligo #1) or a nontargeting (N/T) siRNA. Protein extracts were then immunoblotted for the indicated proteins. Unprocessed original scans of blots are shown Supplementary Fig. 9. Unless otherwise noted, experiments were performed at least three times.
Supplementary Figure 6 Inhibition of KBP degradation induces a gain in mitonchondrial mass similar to that observed during differentiation.
(a) mESCs stably infected with lentiviruses expressing either HA-tagged wild type mouse KBP or HA-tagged mouse KBP(KK/RR) were treated for 12 h with Qc-1. Cells were collected and lysed for immunoblotting as indicated. (b–c) mESCs infected with lentiviruses expressing either HA-tagged wild type mouse KBP or HA-tagged mouse KBP(KK/RR) were either maintained in LIF-containing medium or induced to differentiate for 24 h after LIF withdrawal and exposure to retinoic acid (RA). Where indicated, cells were treated for 12 h with the TDH inhibitor Qc-1. Panel (b) shows the MIPs of representative cells (scale bar, 15 μm). Panel (c) shows the quantification of average mitochondrial count. Error bars indicate ± s.d. n = 108, 17, 56, 129, 66 and 39 cells [KBP – DMSO, Qc-1 and differentiation- and KBP(KK/RR) – DMSO, Qc-1 and differentiation-, respectively]. P value was calculated by two-way ANOVA and multiple comparison. Unprocessed original scans of blots are shown Supplementary Fig. 9. Unless otherwise noted, experiments were performed at least three times.
Supplementary Figure 7 Defects in KBP degradation do not affect mitochondrial dynamics.
(a–f) mESCs were infected with lentiviruses expressing either HA-tagged wild type mouse KBP or HA-tagged mouse KBP(KK/RR) and then transiently transfected with a plasmid expressing mitochondrial-targeted GFP. Panels (a) and (d) show representative 3D reconstructions of the mitochondria. Panels (b) and (e) show the entity of mitochondrial size changes registered in a and d, respectively. Panels (c) and (f) show the quantification of the events of mitochondrial fission and fusion. n = 6 and 7 independent field acquisitions [KBP and KBP(KK/RR), respectively]. Statistics was calculated with unpaired t-test. This experiment was performed once.
Supplementary Figure 8 Generation of Kif1bp−/− mESCs and model of regulation of mitochondrial biogenesis in mESCs.
(a) Generation of Kif1bp−/− mESCs by CRISPR Genome Editing. Schematic representation of Kif1bp genomic locus and gRNA target location. Exon 1 refers to mouse Kif1bp gene in NC_000076.6 Reference GRCm38.p4 C57BL/6J and in NM_028197.2. In the bottom panel, full-length wild-type genomic DNA template and truncated mutant sequences identified by TOPO-TA cloning of Kif1bp PCR from two independent mESCs knockout clones are depicted. Clone #1 sequence and chromatogram revealed an identical indel event at both alleles. Clone #2 sequence and chromatogram identified a 107bp deletion event at both alleles. (b) Protein extracts from wild type mESCs (WT) and Kif1bp-/- mESCs (two different clones) were immunoblotted for the indicated proteins. Unprocessed original scans of blots are shown Supplementary Fig. 9. This experiment was performed three times. (c) The TDH-dependent production of acetyl-CoA at mitochondria of naïve mESCs promotes the GCN5L1-mediated acetylation of KBP on Lysine 501. This event, in turn, allows KBP to be recognized by and degraded via Fbxo15, attenuating mitochondrial biogenesis during self-renewal. This molecular mechanism limits the accumulation of mitochondrial mass in self-renewing mESCs and preserves their optimal fitness. In human, TDH is an expressed pseudogene, a unique case among all metazoans (including chimpanzees) whose genomes have been sequenced to date (Pruitt et al. Nucleic Acids Res. 2007). However, the fact that the degron of KBP is conserved and acetylated in humans, together with the finding that its presence is necessary for KBP binding to Fbxo15, suggests that AcetylCoA is necessary for Fbxo15-mediated degradation of KBP in human cells as well, even if the source of AcetylCoA is different. Altogether, our data suggest that the molecular mechanism we have identified in mouse, with some minor differences, is conserved in humans. (d) Upon differentiation, levels of Fbxo15 and TDH decrease, acetylation of KBP on Lysine 501 is inhibited, and KBP levels accumulate. We hypothesize that this accumulation results in the increase in cellular mitochondrial mass, which contributes to the metabolic switch in differentiating cells.
Supplementary information
Supplementary Information
Supplementary Information (PDF 16774 kb)
Supplementary Table 1
Supplementary Information (XLSX 110 kb)
Supplementary Table 2
Supplementary Information (XLS 8554 kb)
Supplementary Table 3
Supplementary Information (XLS 15775 kb)
Supplementary Table 4
Supplementary Information (XLS 73 kb)
Supplementary Table 5
Supplementary Information (XLSX 38 kb)
Supplementary Table 6
Supplementary Information (XLSX 47 kb)
Rights and permissions
About this article
Cite this article
Donato, V., Bonora, M., Simoneschi, D. et al. The TDH–GCN5L1–Fbxo15–KBP axis limits mitochondrial biogenesis in mouse embryonic stem cells. Nat Cell Biol 19, 341–351 (2017). https://doi.org/10.1038/ncb3491
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb3491
This article is cited by
-
GCN5L1 regulates pulmonary surfactant production by modulating lamellar body biogenesis and trafficking in mouse alveolar epithelial cells
Cellular & Molecular Biology Letters (2023)
-
F-box only protein 9 and its role in cancer
Molecular Biology Reports (2022)
-
Skp2 dictates cell cycle-dependent metabolic oscillation between glycolysis and TCA cycle
Cell Research (2021)
-
CRL4AMBRA1 is a master regulator of D-type cyclins
Nature (2021)
-
Powering stem cell decisions with ubiquitin
Cell Death & Differentiation (2017)