Abstract
Mitochondria drive apoptosis by releasing pro-apoptotic proteins that promote caspase activation in the cytosol. The rhomboid protease PARL, an intramembrane cleaving peptidase in the inner membrane, regulates mitophagy and plays an ill-defined role in apoptosis. Here, we employed PARL-based proteomics to define its substrate spectrum. Our data identified the mitochondrial pro-apoptotic protein Smac (also known as DIABLO) as a PARL substrate. In apoptotic cells, Smac is released into the cytosol and promotes caspase activity by inhibiting inhibitors of apoptosis (IAPs). Intramembrane cleavage of Smac by PARL generates an amino-terminal IAP-binding motif, which is required for its apoptotic activity. Loss of PARL impairs proteolytic maturation of Smac, which fails to bind XIAP. Smac peptidomimetics, downregulation of XIAP or cytosolic expression of cleaved Smac restores apoptosis in PARL-deficient cells. Our results reveal a pro-apoptotic function of PARL and identify PARL-mediated Smac processing and cytochrome c release facilitated by OPA1-dependent cristae remodelling as two independent pro-apoptotic pathways in mitochondria.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Chan, D. C. Fusion and fission: interlinked processes critical for mitochondrial health. Annu. Rev. Genet. 46, 265–287 (2012).
Schrepfer, E. & Scorrano, L. Mitofusins, from mitochondria to metabolism. Mol. Cell 61, 683–694 (2016).
Wai, T. & Langer, T. Mitochondrial dynamics and metabolic regulation. Trends Endocrinol. Metab. 27, 105–117 (2016).
Youle, R. J. & van der Bliek, A. M. Mitochondrial fission, fusion, and stress. Science 337, 1062–1065 (2012).
Quiros, P. M., Langer, T. & Lopez-Otin, C. New roles for mitochondrial proteases in health, ageing and disease. Nat. Rev. Mol. Cell Biol. 16, 345–359 (2015).
Liesa, M. & Shirihai, O. S. Mitochondrial dynamics in the regulation of nutrient utilization and energy expenditure. Cell Metab. 17, 491–506 (2013).
Ehses, S. et al. Regulation of OPA1 processing and mitochondrial fusion by m-AAA protease isoenzymes and OMA1. J. Cell Biol. 187, 1023–1036 (2009).
Griparic, L., Kanazawa, T. & van der Bliek, A. M. Regulation of the mitochondrial dynamin-like protein Opa1 by proteolytic cleavage. J. Cell Biol. 178, 757–764 (2007).
Head, B., Griparic, L., Amiri, M., Gandre-Babbe, S. & van der Bliek, A. M. Inducible proteolytic inactivation of OPA1 mediated by the OMA1 protease in mammalian cells. J. Cell Biol. 187, 959–966 (2009).
Song, Z., Chen, H., Fiket, M., Alexander, C. & Chan, D. C. OPA1 processing controls mitochondrial fusion and is regulated by mRNA splicing, membrane potential, and Yme1L. J. Cell Biol. 178, 749–755 (2007).
Anand, R. et al. The i-AAA protease YME1L and OMA1 cleave OPA1 to balance mitochondrial fusion and fission. J. Cell Biol. 204, 919–929 (2014).
MacVicar, T. & Langer, T. OPA1 processing in cell death and disease—the long and short of it. J. Cell Sci. 129, 2297–2306 (2016).
Cipolat, S. et al. Mitochondrial rhomboid PARL regulates cytochrome c release during apoptosis via OPA1-dependent cristae remodeling. Cell 126, 163–175 (2006).
Frezza, C. et al. OPA1 controls apoptotic cristae remodeling independently from mitochondrial fusion. Cell 126, 177–189 (2006).
Korwitz, A. et al. Loss of OMA1 delays neurodegeneration by preventing stress-induced OPA1 processing in mitochondria. J. Cell Biol. 212, 157–166 (2016).
Jiang, X., Jiang, H., Shen, Z. & Wang, X. Activation of mitochondrial protease OMA1 by Bax and Bak promotes cytochrome c release during apoptosis. Proc. Natl Acad. Sci. USA 111, 14782–14787 (2014).
Pernas, L. & Scorrano, L. Mito-morphosis: mitochondrial fusion, fission, and cristae remodeling as key mediators of cellular function. Annu. Rev. Physiol. 78, 505–531 (2016).
Herlan, M., Vogel, F., Bornhovd, C., Neupert, W. & Reichert, A. S. Processing of Mgm1 by the rhomboid-type protease Pcp1 is required for maintenance of mitochondrial morphology and of mitochondrial DNA. J. Biol. Chem. 278, 27781–27788 (2003).
McQuibban, G. A., Saurya, S. & Freeman, M. Mitochondrial membrane remodelling regulated by a conserved rhomboid protease. Nature 423, 537–541 (2003).
Sesaki, H., Southard, S. M., Hobbs, A. E. & Jensen, R. E. Cells lacking Pcp1p/Ugo2p, a rhomboid-like protease required for Mgm1p processing, lose mtDNA and mitochondrial structure in a Dnm1p-dependent manner, but remain competent for mitochondrial fusion. Biochem. Biophys. Res. Commun. 308, 276–283 (2003).
Duvezin-Caubet, S. et al. OPA1 processing reconstituted in yeast depends on the subunit composition of the m-AAA protease in mitochondria. Mol. Biol. Cell 18, 3582–3590 (2007).
Ishihara, N., Fujita, Y., Oka, T. & Mihara, K. Regulation of mitochondrial morphology through proteolytic cleavage of OPA1. EMBO J. 25, 2966–2977 (2006).
Freeman, M. Rhomboids, signalling and cell biology. Biochem. Soc. Trans. 44, 945–950 (2016).
Jin, S. M. et al. Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. J. Cell Biol. 191, 933–942 (2010).
Sekine, S. et al. Rhomboid protease PARL mediates the mitochondrial membrane potential loss-induced cleavage of PGAM5. J. Biol. Chem. 287, 34635–34645 (2012).
Narendra, D. P. et al. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 8, e1000298 (2010).
Morais, V. A. et al. PINK1 loss-of-function mutations affect mitochondrial complex I activity via NdufA10 ubiquinone uncoupling. Science 344, 203–207 (2014).
Chen, G. et al. A regulatory signaling loop comprising the PGAM5 phosphatase and CK2 controls receptor-mediated mitophagy. Mol. Cell 54, 362–377 (2014).
Wang, Z., Jiang, H., Chen, S., Du, F. & Wang, X. The mitochondrial phosphatase PGAM5 functions at the convergence point of multiple necrotic death pathways. Cell 148, 228–243 (2012).
Shi, G. et al. Functional alteration of PARL contributes to mitochondrial dysregulation in Parkinson’s disease. Hum. Mol. Genet. 20, 1966–1974 (2011).
Heinitz, S., Klein, C. & Djarmati, A. The p.S77N presenilin-associated rhomboid-like protein mutation is not a frequent cause of early-onset Parkinson’s disease. Mov. Disord. 26, 2441–2442 (2011).
Civitarese, A. E. et al. Regulation of skeletal muscle oxidative capacity and insulin signaling by the mitochondrial rhomboid protease PARL. Cell Metab. 11, 412–426 (2010).
Venne, A. S., Vogtle, F. N., Meisinger, C., Sickmann, A. & Zahedi, R. P. Novel highly sensitive, specific, and straightforward strategy for comprehensive N-terminal proteomics reveals unknown substrates of the mitochondrial peptidase Icp55. J. Proteome Res. 12, 3823–3830 (2013).
Ghezzi, D. et al. Mutations in TTC19 cause mitochondrial complex III deficiency and neurological impairment in humans and flies. Nat. Genet. 43, 259–263 (2011).
Mokry, D. Z., Abrahao, J. & Ramos, C. H. Disaggregases, molecular chaperones that resolubilize protein aggregates. An. Acad. Bras. Cienc. 87, 1273–1292 (2015).
Horibata, Y. & Sugimoto, H. StarD7 mediates the intracellular trafficking of phosphatidylcholine to mitochondria. J. Biol. Chem. 285, 7358–7365 (2010).
Du, C., Fang, M., Li, Y., Li, L. & Wang, X. Smac, a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition. Cell 102, 33–42 (2000).
Chai, J. et al. Structural and biochemical basis of apoptotic activation by Smac/DIABLO. Nature 406, 855–862 (2000).
Verhagen, A. M. et al. Identification of DIABLO, a mammalian protein that promotes apoptosis by binding to and antagonizing IAP proteins. Cell 102, 43–53 (2000).
Chao, J. R. et al. Hax1-mediated processing of HtrA2 by Parl allows survival of lymphocytes and neurons. Nature 452, 98–102 (2008).
Ran, F. A. et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154, 1380–1389 (2013).
Mossmann, D., Meisinger, C. & Vogtle, F. N. Processing of mitochondrial presequences. Biochim. Biophys. Acta 1819, 1098–1106 (2012).
Burri, L. et al. Mature DIABLO/Smac is produced by the IMP protease complex on the mitochondrial inner membrane. Mol. Biol. Cell 16, 2926–2933 (2005).
Salvesen, G. S. & Duckett, C. S. IAP proteins: blocking the road to death’s door. Nat. Rev. Mol. Cell Biol. 3, 401–410 (2002).
Holcik, M. & Korneluk, R. G. XIAP, the guardian angel. Nat. Rev. Mol. Cell Biol. 2, 550–556 (2001).
Mannhold, R., Fulda, S. & Carosati, E. IAP antagonists: promising candidates for cancer therapy. Drug Discov. Today 15, 210–219 (2010).
Rettinger, E. et al. SMAC mimetic BV6 enables sensitization of resistant tumor cells but also affects Cytokine-Induced Killer (CIK) cells: a potential challenge for combination therapy. Front. Pediatr. 2, 75 (2014).
Li, W. et al. BV6, an IAP antagonist, activates apoptosis and enhances radiosensitization of non-small cell lung carcinoma in vitro. J. Thorac. Oncol. 6, 1801–1809 (2011).
Eckelman, B. P., Salvesen, G. S. & Scott, F. L. Human inhibitor of apoptosis proteins: why XIAP is the black sheep of the family. EMBO Rep. 7, 988–994 (2006).
Seeger, J. M. et al. Elevated XIAP expression alone does not confer chemoresistance. Br. J. Cancer 102, 1717–1723 (2010).
Okada, H. et al. Generation and characterization of Smac/DIABLO-deficient mice. Mol. Cell Biol. 22, 3509–3517 (2002).
Schug, Z. T. & Gottlieb, E. Cardiolipin acts as a mitochondrial signalling platform to launch apoptosis. Biochim. Biophys. Acta 1788, 2022–2031 (2009).
Jost, P. J. et al. XIAP discriminates between type I and type II FAS-induced apoptosis. Nature 460, 1035–1039 (2009).
Saunders, C. et al. CLPB variants associated with autosomal-recessive mitochondrial disorder with cataract, neutropenia, epilepsy, and methylglutaconic aciduria. Am. J. Hum. Genet. 96, 258–265 (2015).
Wortmann, S. B. et al. CLPB mutations cause 3-methylglutaconic aciduria, progressive brain atrophy, intellectual disability, congenital neutropenia, cataracts, movement disorder. Am. J. Hum. Genet. 96, 245–257 (2015).
Kanabus, M. et al. Bi-allelic CLPB mutations cause cataract, renal cysts, nephrocalcinosis and 3-methylglutaconic aciduria, a novel disorder of mitochondrial protein disaggregation. J. Inherit. Metab. Dis. 38, 211–219 (2015).
Capo-Chichi, J. M. et al. Disruption of CLPB is associated with congenital microcephaly, severe encephalopathy and 3-methylglutaconic aciduria. J. Med. Genet. 52, 303–311 (2015).
Yang, L. et al. Haploinsufficiency for Stard7 is associated with enhanced allergic responses in lung and skin. J. Immunol. 194, 5635–5643 (2015).
Kashkar, H. et al. XIAP targeting sensitizes Hodgkin lymphoma cells for cytolytic T-cell attack. Blood 108, 3434–3440 (2006).
Luciano, P., Geoffroy, S., Brandt, A., Hernandez, J. F. & Geli, V. Functional cooperation of the mitochondrial processing peptidase subunits. J. Mol. Biol. 272, 213–225 (1997).
Venne, A. S. et al. An improved workflow for quantitative N-terminal charge-based fractional diagonal chromatography (ChaFRADIC) to study proteolytic events in Arabidopsis thaliana. Proteomics 15, 2458–2469 (2015).
Burkhart, J. M., Schumbrutzki, C., Wortelkamp, S., Sickmann, A. & Zahedi, R. P. Systematic and quantitative comparison of digest efficiency and specificity reveals the impact of trypsin quality on MS-based proteomics. J. Proteomics 75, 1454–1462 (2012).
Thingholm, T. E., Palmisano, G., Kjeldsen, F. & Larsen, M. R. Undesirable charge-enhancement of isobaric tagged phosphopeptides leads to reduced identification efficiency. J. Proteome Res. 9, 4045–4052 (2010).
Rappsilber, J., Ishihama, Y. & Mann, M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 75, 663–670 (2003).
Cox, J. & Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 (2008).
Cox, J. et al. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 (2011).
Tusher, V. G., Tibshirani, R. & Chu, G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl Acad. Sci. USA 98, 5116–5121 (2001).
Hunter, J. D. Matplotlib: a 2D graphics environment. Comput. Sci. Eng. 9, 90–95 (2007).
Acknowledgements
We thank E. Barth for technical support in electron microscopy and other laboratory members for their support. This work was supported a Japan Society for the Promotion of Science (JSPS) Fellowship for Research Abroad to S.S. and a Reinhart Koselleck grant of the Deutsche Forschungsgemeinschaft to T.L. R.P.Z. and A.S.V. thank the Ministerium für Innovation, Wissenschaft und Forschung des Landes Nordrhein-Westfalen, the Senatsverwaltung für Wirtschaft, Technologie und Forschung des Landes Berlin and the Bundesministerium für Bildung und Forschung for financial support. We thank B. de Strooper (VIB, Leuven) for the Parl−/− MEFs and E. Shoubridge (McGill University, Montreal) for the pBABE-puro vector.
Author information
Authors and Affiliations
Contributions
S.S. and T.L. designed the research. S.S. and K.U.F. performed the experiments. H.N. and M.K. performed the mass spectrometric analysis for differential proteomics. A.S.V. and R.P.Z. performed the ChaFRADIC analysis. H.K. provided essential tools and reagents. S.S. and T.L. wrote the manuscript with the comments of the other authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 PARL-based proteomics.
(a) Proteomic strategies for the identification of PARL substrate proteins in mitochondria. Left panel; determination of N-terminal peptides of mitochondrial proteins quantitative affinity purification proteomics. (b) iTRAQ based ratios were log2 transformed and biological replicates were plotted against each other. Reproducibility was determined by Pearson correlation coefficient: r > 0.4 (after filtering proteins with higher standard deviations than 99% of the complete population). Proteins that were found with extreme ratios (>0.99 quantile) are highlighted by their gene name. (c) Isolation of mitochondria from wild type (WT) and PARL−/− HEK293 cells by Percoll gradient centrifugation. Fraction 3 was collected and analysed by ChaFRADIC (Supplementary Table 1). (d) SDS-PAGE and immunoblot analysis of cellular fractions of wild type (WT) and PARL−/− HEK293 cells. p, precursor; i, intermediate; m, mature form. Unprocessed original scans of blots are shown in Supplementary Fig. 9.
Supplementary Figure 2 Characterization of Smac/DIABLO in PARL deficient cells.
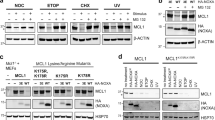
(a) Fractionation of wild type (WT) and PARL−/− HEK293 cells into post nuclear (PNS), mitochondrial (mito), microsomal and cytosolic fractions. Samples were analysed by SDS-PAGE and immunoblotting. p, precursor; i, intermediate; m, mature. (b) siRNA-mediated downregulation of Smac/DIABLO in wild type (WT) and PARL−/− HEK293 cells for 72 h (or with control siRNA). Cell lysates were analysed by SDS-PAGE and immunoblotting. p, precursor; i, intermediate; m, mature. (c) Transient expression of Smac/DIABLOHA in wild type (WT) and PARL−/− (KO) HEK293 cells for 24 h (reagent 1, GeneJuice; reagent 2, Lipofectamine 2000). Cell lysates were analysed by SDS-PAGE and immunoblotting. p, precursor; i, intermediate; m, mature. (d) Quantification of different Smac/DIABLO forms in wild type (WT) and PARL−/− HEK293 cells expressing PARLFLAG or PARLS277A−FLAG when indicated. Left panel; cell lysates were analysed by SDS-PAGE and immunoblotting. Right panel; quantification of fluorescence intensities using the infrared Odyssey System (n = 3 independent experiments; mean values ± SD). p, precursor; i, intermediate; m, mature. (e–g) SDS-PAGE and immunoblot analysis of cell extracts of wild type (WT) and PARL-deficient (KO) MEFs (e), HeLa cells (f), HCT116 (g) cells. Different clones of PARL−/− HeLa and HCT116 cells were analysed. a–e, OPA1 forms, p, precursor; i, intermediate; m, mature form. ∗unspecific cross-reaction. Unprocessed original scans of blots are shown in Supplementary Fig. 9.
Supplementary Figure 3 Processing of Smac/DIABLO.
(a) Hydropathy plot of the N-terminal 90 amino acids of Smac/DIABLO. The red bar indicates a predicted transmembrane domain (W42–A60). (b) Accumulation of the intermediate form of Smac/DIABLO upon downregulation of MPP in wild type (WT) and PARL−/− HEK293 cells. siRNAs directed against α- and β-Mpp (or control siRNA) were transfected for 72 h. Cell lysates were analysed by SDS-PAGE and immunoblotting. p, precursor; i, intermediate; m, mature form. ∗unspecific cross-reaction. (c) siRNA-mediated downregulation of MPP in wild type (WT) and PARL−/− HEK293 cells expressing Smac/DIABLOHA (for 24 h) when indicated. Cell lysates were analysed by SDS-PAGE and immunoblotting. p, precursor; i, intermediate; m, mature form. ∗unspecific cross-reaction. (d) Recombinant Smac/DIABLOHA (synthesized in rabbit reticulocyte lysate) was incubated for 1 h at 30 °C with mitochondrial extracts of wild type (WT) and PARL−/− HEK293 cells in the absence or presence of CCCP, o-phenanthroline, DCI, PMSF or cOmplete inhibitor mix. Samples were analysed by SDS-PAGE and immunoblotting. p, precursor; i, intermediate; m, mature form. (e,f) Downregulation of (e) IMMPL1/2L or (f) HTRA2/Omi in PARL wild type (WT) and PARL−/− HEK293 cells for 72 h (or with control siRNA). Cell lysates were analysed by SDS-PAGE and immunoblotting. p, precursor; i, intermediate; m, mature form. ∗unspecific cross-reaction. Unprocessed original scans of blots are shown in Supplementary Fig. 9.
Supplementary Figure 4 Increased apoptotic resistance of PARL-deficient MEFs and HeLa cells.
(a) Total RNA was extracted from wild type (WT) and PARL−/− HeLa or HCT116 cells complemented with PARLFLAG or PARLS277A−FLAG and subjected to RT and real-time PCR analysis of human PARL mRNA. (n = 3 independent experiments; mean values ± SD). (b) Wild type (WT) and Parl−/− MEFs were treated for 8 h with staurosporine (STS, 1 μM), actinomycin D (Act D, 1 μM), or a combination of TNFα (10 ng ml−1) and CHX (10 μg ml−1). Cell lysates were analysed by SDS-PAGE and immunoblotting. p, precursor; i, intermediate; m, mature form. ∗unspecific cross-reaction. (c) Quantification of PARP cleavage of experiments in b (n = 3 independent experiments; mean values ± s.d.). P values were calculated by one-way ANOVA and indicated in figure. A P value of <0.05 was considered statistically significant. (d) Viability of wild type and Parl−/− MEFs (complemented with PARLFLAG when indicated) after incubation for 8 h with staurosporine (STS, 0.1 μM) (n = 3 independent experiments; mean values ± s.d.). Cell viability was assessed monitoring cellular ATP levels. P values were calculated by one-way ANOVA and indicated in figure. A P value of <0.05 was considered statistically significant. (e) Relative confluency of wild type (WT) and PARL−/− HeLa cells in the presence of actinomycin D (Act D, 1 μM). Images of the cells after 16 h Act D treatment are shown in the lower panel. Scale bars, 100 μm. Unprocessed original scans of blots are shown in Supplementary Fig. 9. Statistical source data can be found in Supplementary Table 3.
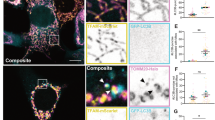
Supplementary Figure 5 The loss of PARL does not affect mitochondrial morphology.
(a) Immunofluorescence analysis of wild type (WT) and PARL−/− HEK293 cells using antibodies directed against ATP5β (green) and TOMM20 (red), and examined by confocal microscopy. Higher-magnification views of the boxed areas were shown. Scale bars, 10 μm or 2 μm (magnified image). (b,c) Immunofluorescence analysis of wild type (WT) and PARL−/− HCT116 cells (b), and HeLa cells (c) using antibodies directed against TOMM20 (green) and ATP5β (red), and examined by confocal microscopy. Higher magnifications of the boxed areas are shown. Scale bars, (b), 10 μm or 5 μm (magnified image); (c) 20 μm or 5 μm (magnified image). (d) Transmission electron microscopic analysis of wild type (WT) and PARL−/− HeLa cells (complemented with PARLFLAG when indicated). Scale bars, 1 μm. (e) BAX oligomerization in PARL-deficient HCT116 cells. Wild type (WT) and PARL−/− HCT116 cells were incubated with TNFα (20 ng ml−1) and CHX (10 μg ml−1) for 6 h. Mitochondria were isolated and analysed by BN-PAGE or SDS-PAGE, followed by immunoblotting with the indicated antibodies. Unprocessed original scans of blots are shown in Supplementary Fig. 9.
Supplementary Figure 6 Quantification of XIAP mRNA and protein in human and mouse cell lines.
(a) Immunoblot analysis of cell lysates of wild type (WT) or PARL−/− cells using the indicated antibodies. ∗unspecific cross-reaction. (b) Quantification of experiments in c (n = 3 independent experiments; mean values ± s.d.). P values were calculated by one-way ANOVA and indicated in figure. A P value of <0.05 was considered statistically significant. Statistical source data can be found in Supplementary Table 3. Unprocessed original scans of blots are shown in Supplementary Fig. 9.
Supplementary Figure 7 Depletion of XIAP restores apoptosis in PARL-deficient cells.
(a) After transfection with Xiap (or with control) siRNA for 48 h, wild type (WT) and PARL−/− (KO) HCT116 cells were incubated with TNFα (20 ng ml−1) and CHX (10 μg ml−1). Cell lysates were analysed by SDS-PAGE and immunoblotting. p, precursor; i, intermediate; m, mature form. ∗unspecific cross-reaction. (b) Quantification of PARP cleavage in experiments described in a (n = 3 independent experiments); mean values ± s.d.). P values were calculated by one-way ANOVA and indicated in figure. A P value of <0.05 was considered statistically significant. Unprocessed original scans of blots are shown in Supplementary Fig. 9. Statistical source data can be found in Supplementary Table 3.
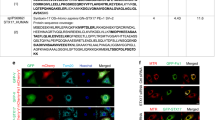
Supplementary Figure 8 Cytosolic, mature Smac/DIABLO is sufficient to drive apoptosis in PARL−/− cells.
(a, left) Wild type (WT) and PARL−/− HCT116 cells were incubated with TNFα (20 ng ml−1) and CHX (10 μg ml−1) for 6 h in the absence or presence of the Smac/DIABLO mimetic BV6 (1 μM). (a, right) Wild type (WT) and PARL−/− HeLa cells were incubated with staurosporine (STS, 0.1 μM) for 6 h in the absence or presence of the Smac/DIABLO mimetic BV6 (2.5 μM). p, precursor; i, intermediate; m, mature form. ∗unspecific cross-reaction. Unprocessed original scans of blots are shown in Supplementary Fig. 9. (b) Smac/DIABLO-deficient HeLa cells were generated by CRISPR/Cas9-mediated genome editing. Variants of Smac/DIABLO predicted to be expressed are shown. The red asterisk indicates the CRISPR/Cas9 targeted site. (c) Domain organization of Ubiquitin-Smac/DIABLO (Δ55)-DsRed and Ubiquitin-Smac/DIABLO (Δ59)-DsRed. (d) Two pathways for the release of pro-apoptotic proteins from mitochondria. BAX/BAK-induced apoptosis requires PARL-mediated processing and cytosolic translocation of Smac/DIABLO and the release of cytochrome c, which is facilitated by OPA1-dependent cristae remodeling and cardiolipin oxidation.
Supplementary information
Supplementary Information
Supplementary Information (PDF 19513 kb)
Supplementary Table 1
Supplementary Information (XLSX 1209 kb)
Supplementary Table 2
Supplementary Information (XLSX 2051 kb)
Supplementary Table 3
Supplementary Information (XLSX 106 kb)
Supplementary Table 4
Supplementary Information (XLSX 14 kb)
Supplementary Table 5
Supplementary Information (XLSX 31 kb)
Act D treated HeLa cells (related to Supplementary Fig. 4e).
Relative confluency of wild type (WT) HeLa cells in the presence of actinomycin D (Act D, 1 μM). Images were taken every 5 min for 16 h. (MOV 7923 kb)
Act D treated PARL−/− HeLa cells (related to Supplementary Fig. 4e).
Relative confluency of PARL−/− (KO) HeLa cells in the presence of actinomycin D (Act D, 1 μM). Images were taken every 5 min for 16 h. (MOV 10865 kb)
Rights and permissions
About this article
Cite this article
Saita, S., Nolte, H., Fiedler, K. et al. PARL mediates Smac proteolytic maturation in mitochondria to promote apoptosis. Nat Cell Biol 19, 318–328 (2017). https://doi.org/10.1038/ncb3488
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb3488
This article is cited by
-
The mitochondrial protease PARL is required for spermatogenesis
Communications Biology (2024)
-
Sub-lethal signals in the mitochondrial apoptosis apparatus: pernicious by-product or physiological event?
Cell Death & Differentiation (2023)
-
Structural basis of impaired disaggregase function in the oxidation-sensitive SKD3 mutant causing 3-methylglutaconic aciduria
Nature Communications (2023)
-
Mitochondria regulate intracellular coenzyme Q transport and ferroptotic resistance via STARD7
Nature Cell Biology (2023)
-
Protease-independent control of parthanatos by HtrA2/Omi
Cellular and Molecular Life Sciences (2023)