Abstract
Chromosome missegregation into a micronucleus can cause complex and localized genomic rearrangements1,2 known as chromothripsis3, but the underlying mechanisms remain unresolved. Here we developed an inducible Y centromere-selective inactivation strategy by exploiting a CENP-A/histone H3 chimaera to directly examine the fate of missegregated chromosomes in otherwise diploid human cells. Using this approach, we identified a temporal cascade of events that are initiated following centromere inactivation involving chromosome missegregation, fragmentation, and re-ligation that span three consecutive cell cycles. Following centromere inactivation, a micronucleus harbouring the Y chromosome is formed in the first cell cycle. Chromosome shattering, producing up to 53 dispersed fragments from a single chromosome, is triggered by premature micronuclear condensation prior to or during mitotic entry of the second cycle. Lastly, canonical non-homologous end joining (NHEJ), but not homology-dependent repair, is shown to facilitate re-ligation of chromosomal fragments in the third cycle. Thus, initial errors in cell division can provoke further genomic instability through fragmentation of micronuclear DNAs coupled to NHEJ-mediated reassembly in the subsequent interphase.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Tan, E. H. et al. Catastrophic chromosomal restructuring during genome elimination in plants. eLife 4, e06516 (2015).
Zhang, C.-Z. et al. Chromothripsis from DNA damage in micronuclei. Nature 522, 179–184 (2015).
Stephens, P. J. et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell 144, 27–40 (2011).
Janssen, A. et al. Chromosome segregation errors as a cause of DNA damage and structural chromosome aberrations. Science 333, 1895–1898 (2011).
Rausch, T. et al. Genome sequencing of pediatric medulloblastoma links catastrophic DNA rearrangements with TP53 mutations. Cell 148, 59–71 (2012).
Maciejowski, J., Li, Y., Bosco, N., Campbell, P. J. & de Lange, T. Chromothripsis and kataegis induced by telomere crisis. Cell 163, 1641–1654 (2015).
Mardin, B. R. et al. A cell-based model system links chromothripsis with hyperploidy. Mol. Syst. Biol. 11, 828–828 (2015).
Sabatinos, S. A., Ranatunga, N. S., Yuan, J.-P., Green, M. D. & Forsburg, S. L. Replication stress in early S phase generates apparent micronuclei and chromosome rearrangement in fission yeast. Mol. Biol. Cell 26, 3439–3450 (2015).
Crasta, K. et al. DNA breaks and chromosome pulverization from errors in mitosis. Nature 482, 53–58 (2012).
Holland, A. J. & Cleveland, D. W. Chromoanagenesis and cancer: mechanisms and consequences of localized, complex chromosomal rearrangements. Nat. Med. 18, 1630–1638 (2012).
Kato, H. & Sandberg, A. A. Chromosome pulverization in human cells with micronuclei. J. Natl Cancer Inst. 40, 165–179 (1968).
Orth, J. D., Loewer, A., Lahav, G. & Mitchison, T. J. Prolonged mitotic arrest triggers partial activation of apoptosis, resulting in DNA damage and p53 induction. Mol. Biol. Cell 23, 567–576 (2012).
Ganem, N. J. & Pellman, D. Linking abnormal mitosis to the acquisition of DNA damage. J. Cell Biol. 199, 871–881 (2012).
Dalton, W. B. et al. Human cancer cells commonly acquire DNA damage during mitotic arrest. Cancer Res. 67, 11487–11492 (2007).
Hayashi, M. T., Cesare, A. J., Fitzpatrick, J. A. J., Lazzerini-Denchi, E. & Karlseder, J. A telomere-dependent DNA damage checkpoint induced by prolonged mitotic arrest. Nat. Struct. Mol. Biol. 19, 387–394 (2012).
Klare, K. et al. CENP-C is a blueprint for constitutive centromere-associated network assembly within human kinetochores. J. Cell Biol. 210, 11–22 (2015).
Carroll, C. W., Milks, K. J. & Straight, A. F. Dual recognition of CENP-A nucleosomes is required for centromere assembly. J. Cell Biol. 189, 1143–1155 (2010).
Fachinetti, D. et al. DNA sequence-specific binding of CENP-B enhances the fidelity of human centromere function. Dev. Cell 33, 314–327 (2015).
Fachinetti, D. et al. A two-step mechanism for epigenetic specification of centromere identity and function. Nat. Cell Biol. 15, 1–13 (2013).
Logsdon, G. A. et al. Both tails and the centromere targeting domain of CENP-A are required for centromere establishment. J. Cell Biol. 208, 521–531 (2015).
Guse, A., Carroll, C. W., Moree, B., Fuller, C. J. & Straight, A. F. In vitro centromere and kinetochore assembly on defined chromatin templates. Nature 477, 354–358 (2011).
Masumoto, H., Masukata, H., Muro, Y., Nozaki, N. & Okazaki, T. A human centromere antigen (CENP-B) interacts with a short specific sequence in alphoid DNA, a human centromeric satellite. J. Cell Biol. 109, 1963–1973 (1989).
Earnshaw, W. C. et al. Molecular cloning of cDNA for CENP-B, the major human centromere autoantigen. J. Cell Biol. 104, 817–829 (1987).
Thompson, S. L. & Compton, D. A. Proliferation of aneuploid human cells is limited by a p53-dependent mechanism. J. Cell Biol. 188, 369–381 (2010).
Hinchcliffe, E. H. et al. Chromosome missegregation during anaphase triggers p53 cell cycle arrest through histone H3.3 Ser31 phosphorylation. Nat. Cell Biol. 18, 668–675 (2016).
Holland, A. J., Fachinetti, D., Han, J. S. & Cleveland, D. W. Inducible, reversible system for the rapid and complete degradation of proteins in mammalian cells. Proc. Natl Acad. Sci. USA 109, E3350–E3357 (2012).
Nishimura, K., Fukagawa, T., Takisawa, H., Kakimoto, T. & Kanemaki, M. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat. Methods 6, 917–922 (2009).
Jansen, L. E. T., Black, B. E., Foltz, D. R. & Cleveland, D. W. Propagation of centromeric chromatin requires exit from mitosis. J. Cell Biol. 176, 795–805 (2007).
Leach, N. T. & Jackson-Cook, C. Micronuclei with multiple copies of the X chromosome: do chromosomes replicate in micronuclei? Mutat. Res. 554, 89–94 (2004).
Shimizu, N., Kanda, T. & Wahl, G. M. Selective capture of acentric fragments by micronuclei provides a rapid method for purifying extrachromosomally amplified DNA. Nat. Genet. 12, 65–71 (1996).
Hatch, E. M., Fischer, A. H., Deerinck, T. J. & Hetzer, M. W. Catastrophic nuclear envelope collapse in cancer cell micronuclei. Cell 154, 47–60 (2013).
Terradas, M., Martín, M., Tusell, L. & Genescà, A. DNA lesions sequestered in micronuclei induce a local defective-damage response. DNA Rep. 8, 1225–1234 (2009).
Johnson, R. T. & Rao, P. N. Mammalian cell fusion: induction of premature chromosome condensation in interphase nuclei. Nature 226, 717–722 (1970).
Gavet, O., Gavet, O. & Pines, J. Activation of cyclin B1-Cdk1 synchronizes events in the nucleus and the cytoplasm at mitosis. J. Cell Biol. 189, 247–259 (2010).
Ceccaldi, R., Rondinelli, B. & D’Andrea, A. D. Repair pathway choices and consequences at the double-strand break. Trends Cell Biol. 26, 52–64 (2016).
Malhotra, A. et al. Breakpoint profiling of 64 cancer genomes reveals numerous complex rearrangements spawned by homology-independent mechanisms. Genome Res. 23, 762–776 (2013).
Kloosterman, W. P. et al. Constitutional chromothripsis rearrangements involve clustered double-stranded DNA breaks and nonhomologous repair mechanisms. Cell Rep. 1, 648–655 (2012).
Campbell, P. J. et al. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat. Genet. 40, 722–729 (2008).
Skaletsky, H. et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature 423, 825–837 (2003).
Ghezraoui, H. et al. Chromosomal translocations in human cells are generated by canonical nonhomologous end-joining. Mol. Cell 55, 829–842 (2014).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17, 10–12 (2011).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Acknowledgements
We thank K. Jepsen and the UC San Diego IGM Genomics Center (MCC P30 CA023100) for DNA library preparation and sequencing, A. Shiau, S. Dowdy, E. Hatch, M. Hetzer, X. Wu and T. Pyntikova (Whitehead Institute, USA) for providing reagents, D. Jenkins, I. Goyal and Y. Sun for technical assistance, and the UC San Diego School of Medicine Microscopy Core (NINDS P30 NS047101) for shared use of equipment. This work was funded by a grant from the NIH (GM029513) to D.W.C., who receives salary support from the Ludwig Institute for Cancer Research. D.C.P. is supported by the Howard Hughes Medical Institute and NIH (HG007852). P.L. was supported by a Cancer Cell Biology Training Grant from the NCI (5T32CA067754-18) and a Postdoctoral Fellowship from the Hope Funds for Cancer Research (HFCR-14-06-06).
Author information
Authors and Affiliations
Contributions
P.L. and D.W.C. conceived the project, designed the experiments, and wrote the manuscript. P.L. conducted the experiments and analysed the data. D.F. constructed the parental AID-tagged CENP-A cell line and provided key experimental input. P.L. and D.H.K. performed purification of micronuclei. O.S. assisted with FISH experiments. L.S.T., H.S. and D.C.P. analysed the sequencing data. All authors contributed comments on the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Construction of human DLD-1 cells with auxin-degradable CENP-AAID and a doxycycline-inducible CENP-AC−H3 rescue that is capable of maintaining centromere identity and function.
(a) Amino acid sequence of wild-type CENP-A (WT) and the carboxy-terminal tail chimera (CH-3) swapped with the corresponding tail of histone H3. Schematic not drawn to scale. CATD; centromere targeting domain. (b) Schematic for the construction of DLD-1 cell lines used in all experiments. (c) Unfixed DLD-1 CENP-AEYFP−AID cells stably expressing H2B-mRFP were imaged 2d after IAA addition. Scale bar, 5 μm. (d) DLD-1 cells as in (b) were treated with combinations of dox and IAA for 24 h and whole-cell extracts were analyzed by immunoblotting for CENP-A. The predicted molecular weight of CENP-A fused to an EYFP-AID tag is ∼66 kDa. (e) Representative immunofluorescent images of engineered DLD-1 cells treated with combinations of dox and IAA for 24 h. Both CENP-AWT and CENP-AC−H3 rescues correctly localized to centromeres. Enlarged images of CENP-A staining following dox/IAA addition is shown below. ACA; anti-centromere antibodies. Scale bar, 5 μm. (f) Dox-inducible CENP-A is expressed at low basal levels without supplemented doxycycline, allowing for the simultaneous addition of dox and IAA without epigenetic loss of centromere identity. (g) CENP-AC−H3-rescued cells are capable of sustaining long-term clonal growth and viability using a 2-week colony formation assay. Data were normalized to untreated cells and represent the mean ± SEM of n = 3 independent experiments each performed in biological triplicate. Asterisks indicate significance by two-tailed Student’s t-test compared to untreated cells. ∗∗P = 0.0019, NS = not significant. (h) Quantification of DLD-1 CENP-AC−H3 cell growth rate with or without dox/IAA over a 9d period performed in biological triplicate. Line represents linear regression analysis. (i) Estimated doubling time calculated from h. (j) 5d CENP-AC−H3-rescued cells were subjected to propidium iodine staining followed by flow cytometry analysis for DNA content with and without 6 h treatment with 100 ng ml−1 nocodazole. Source data for g and h have been provided in Supplementary Table 1.
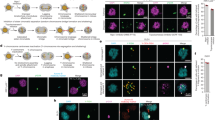
Supplementary Figure 2 Induced Y centromere inactivation provokes Y chromosome missegregation into micronuclei.
(a) DLD-1 cells were rescued with CENP-AWT or CENP-AC−H3 for 5d and the percentage of micronucleated cells were quantified by DAPI staining. Data represent the mean ± SEM of n = 3 independent experiments (1,453–1,945 cells per condition). P-values indicate significance by two-tailed Student’s t-test compared to untreated cells. (b) DLD-1 cells were rescued with CENP-AWT and CENP-AC−H3 for 5d and micronuclei were quantified for the percentage harboring centromere Y or centromere 4 signal(s). Data represent the mean ± SEM of n = 3 independent experiments (380–754 micronuclei) or the mean of 2 independent experiments (CENP-AWT, dox/IAA; 290 micronuclei). P-values indicate significance by two-tailed Student’s t-test compared as denoted. (c) Comparison between the frequency of cells with the specified chromosome in micronuclei when treated as indicated by extrapolating the percentage of micronucleated cells and the percentage of micronuclei containing either chromosome Y or 4. (d) Quantification of the number of Y centromere foci observed in spontaneously-derived or induced micronuclei following 5d CENP-AC−H3 rescue. Data represent the mean ± SEM of n = 65 (WT, -dox/IAA), 76 (WT, + dox/IAA), 67 (C-H3, − dox/IAA) or 102 (C-H3, + dox/IAA) micronuclei. (e) Comparison between the number of Y centromere foci per micronucleus and micronuclear diameter from 5d CENP-AC−H3-rescued cells. Data were compiled from n = 150 micronuclei pooled from 3 independent experiments, and means are indicated by the line. R2-value represents correlation of size and foci number by linear regression analysis. (f) Summary of cellular characteristics comparing untreated (CENP-AEYFP−AID) cells with dox/IAA-treated (CENP-AC−H3) cells. Source data for a and b have been provided in Supplementary Table 1.
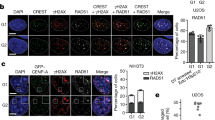
Supplementary Figure 3 Induced Y chromosome micronuclei share common features of spontaneously derived micronuclei including micronuclear envelope disruption and the acquisition of DNA damage.
(a) DLD-1 CENP-AC−H3 cells stably expressing 2xRFP-NLS treated with or without 5d dox/IAA (experimentally versus spontaneously derived micronuclei, respectively) were fixed and DAPI-stained. Representative images from dox/IAA-treated cells and quantifications for micronuclear RFP compartmentalization are shown on the left. Data on the right panel represent the mean of 2 independent experiments (166–294 total micronuclei). Scale bar, 5 μm. (b) DLD-1 CENP-AC−H3 cells treated with 5d dox/IAA were immunostained for the DNA damage marker γH2AX and nuclear envelopes with Lamin B1. Representative images (scale bar, 5 μm) of micronuclei without and with varying degrees of detectable DNA damage signals are shown. (c) γH2AX fluorescent signal intensities from b were measured from 200 micronuclei (pooled from 3 independent experiments) and individually plotted. a.u., arbitrary units. Source data for a and c have been provided in Supplementary Table 1.
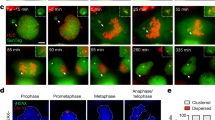
Supplementary Figure 4 Characterization of chromosome fragmentation events and induction of premature chromosome condensation using calyculin A.
(a) Representative example of interphase cells hybridized to Y chromosome paint (green) and Y centromere (red) FISH probes following 3d CENP-AC−H3 rescue. Scale bar, 10 μm. (b) Additional example of Y chromosome fragmentation event derived from 3d CENP-AC−H3-rescued cells. Scale bar, 10 μm. (c) Centromere and fragment counts from Fig. 3f, g were cross-plotted per mitotic shattering event. Red line indicates linear regression analysis. (d) Representative image of DAPI-stained, metaphase-like spreads induced by 1 h treatment with calyculin A, showing examples for G1-, S-, and G2-phase spreads. G1-phase chromosomes appear as single chromatids, S-phase appears as highly pulverized and abnormal nuclei (and excluded from quantitative analyses), and G2-phase appears as normal mitotic chromosomes with two distinguishable sister chromatids. Scale bar, 25 μm. (e) DLD-1 cells treated with 1 μM of the CDK4/6 inhibitor PD-0332991 (also known as Palbociclib) or 10 μM of the CDK1 inhibitor RO-3306 for 24 h were subjected to propidium iodine staining followed by flow cytometry analysis for DNA content. (f) Experimental schematic for panels shown in Fig. 4b, d, e, f, g, h.
Supplementary Figure 5 Paired-end sequencing information for each source of DNA.
(a) Base pair sizes (mean ± SD, n = 1,000 reads) of sequencing fragments for each sample. (b) Percentage of sequencing reads in which both ends of a pair mapped to the reference genome following removal of duplicate and mitochondrial reads. (c) The concentration of discordant sequencing reads for each chromosome follows a second order reaction and rises as the square of the concentration of total sequencing reads (see Methods). Each dot represents a single chromosome from three independent genomic or micronuclear DNA samples, and the green dot indicates the Y chromosome. The curved line shows a predictive model of discordant pairs that is described under the Methods section. Source data for a and b have been provided in Supplementary Table 1.
Supplementary information
Supplementary Information
Supplementary Information (PDF 3075 kb)
Supplementary Table 1
Supplementary Information (XLSX 69 kb)
Rights and permissions
About this article
Cite this article
Ly, P., Teitz, L., Kim, D. et al. Selective Y centromere inactivation triggers chromosome shattering in micronuclei and repair by non-homologous end joining. Nat Cell Biol 19, 68–75 (2017). https://doi.org/10.1038/ncb3450
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb3450
This article is cited by
-
Mosaic derivative chromosomes at chorionic villi (CV) sampling are expression of genomic instability and precursors of cryptic disease-causing rearrangements: report of further four cases
Molecular Cytogenetics (2024)
-
Scrambling the genome in cancer: causes and consequences of complex chromosome rearrangements
Nature Reviews Genetics (2024)
-
The two sides of chromosomal instability: drivers and brakes in cancer
Signal Transduction and Targeted Therapy (2024)
-
Genetic determinants of micronucleus formation in vivo
Nature (2024)
-
Genomic instability in individuals with sex determination defects and germ cell cancer
Cell Death Discovery (2023)