Abstract
Melanoma originates in the epidermis and becomes metastatic after invasion into the dermis. Prior interactions between melanoma cells and dermis are poorly studied. Here, we show that melanoma cells directly affect the formation of the dermal tumour niche by microRNA trafficking before invasion. Melanocytes, cells of melanoma origin, are specialized in releasing pigment vesicles, termed melanosomes. In melanoma in situ, we found melanosome markers in distal fibroblasts before melanoma invasion. The melanosomes carry microRNAs into primary fibroblasts triggering changes, including increased proliferation, migration and pro-inflammatory gene expression, all known features of cancer-associated fibroblasts (CAFs). Specifically, melanosomal microRNA-211 directly targets IGF2R and leads to MAPK signalling activation, which reciprocally encourages melanoma growth. Melanosome release inhibitor prevented CAF formation. Since the first interaction of melanoma cells with blood vessels occurs in the dermis, our data suggest an opportunity to block melanoma invasion by preventing the formation of the dermal tumour niche.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Accession codes
References
Bell, R. E. & Levy, C. The three M’s: melanoma, microphthalmia-associated transcription factor and microRNA. Pigment Cell Melanoma Res. 24, 1088–1106 (2011).
Raposo, G. & Marks, M. S. Melanosomes - dark organelles enlighten endosomal membrane transport. Nat. Rev. Mol. Cell Biol. 8, 786–797 (2007).
Watabe, H., Kushimoto, T., Valencia, J. C. & Hearing, V. J. in Current Protocols in Cell Biology (eds Bonifacino, J. S. et al.) Ch. 3, Unit 3, 14 (2005).
Joshi, P. G. Melanocyte–keratinocyte interaction induces calcium signalling and melanin transfer to keratinocytes. Pigment Cell Res. 20, 380–384 (2007).
Yamaguchi, Y. & Hearing, V. J. Physiological factors that regulate skin pigmentation. Biofactors 35, 193–199 (2009).
Basrur, V. et al. Proteomic analysis of early melanosomes: identification of novel melanosomal proteins. J. Proteome Res. 2, 69–79 (2003).
Boissy, R. E. Melanosome transfer to and translocation in the keratinocyte. Exp. Dermatol. 12, 5–12 (2003).
Lazova, R. & Pawelek, J. M. Why do melanomas get so dark? Exp. Dermatol. 18, 934–938 (2009).
Chen, K. G. et al. Melanosomal sequestration of cytotoxic drugs contributes to the intractability of malignant melanomas. Proc. Natl Acad. Sci. USA 103, 9903–9907 (2006).
Bartel, D. P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297 (2004).
Lu, J. et al. MicroRNA expression profiles classify human cancers. Nature 435, 834–838 (2005).
Levy, C. et al. Intronic miR-211 assumes the tumor suppressive function of its host gene in melanoma. Mol. Cell 40, 841–849 (2010).
Asangani, I. et al. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene 27, 2128–2136 (2008).
Levy, C. et al. Lineage-specific transcriptional regulation of DICER by MITF in melanocytes. Cell 141, 994–1005 (2010).
Bedrosian, I. et al. Incidence of sentinel node metastasis in patients with thin primary melanoma (< or = 1 mm) with vertical growth phase. Ann. Surg. Oncol. 7, 262–267 (2000).
Sorrell, J. M. & Caplan, A. I. Fibroblast heterogeneity: more than skin deep. J. Cell Sci. 117, 667–675 (2004).
Bhowmick, N. A., Neilson, E. G. & Moses, H. L. Stromal fibroblasts in cancer initiation and progression. Nature 432, 332–337 (2004).
Kalluri, R. & Zeisberg, M. Fibroblasts in cancer. Nat. Rev. Cancer 6, 392–401 (2006).
Servais, C. & Erez, N. From sentinel cells to inflammatory culprits: cancer-associated fibroblasts in tumour-related inflammation. J. Pathol. 229, 198–207 (2013).
Orimo, A. et al. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell 121, 335–348 (2005).
Tlsty, T. D. & Hein, P. W. Know thy neighbor: stromal cells can contribute oncogenic signals. Curr. Opin. Genet. Dev. 11, 54–59 (2001).
Cornil, I. et al. Fibroblast cell interactions with human melanoma cells affect tumor cell growth as a function of tumor progression. Proc. Natl Acad. Sci. USA 88, 6028–6032 (1991).
Erez, N., Truitt, M., Olson, P., Arron, S. T. & Hanahan, D. Cancer-associated fibroblasts are activated in incipient neoplasia to orchestrate tumor-promoting inflammation in an NF-κB-dependent manner. Cancer Cell 17, 135–147 (2010).
Quante, M. et al. Bone marrow-derived myofibroblasts contribute to the mesenchymal stem cell niche and promote tumor growth. Cancer Cell 19, 257–272 (2011).
Sharon, Y. et al. Tumor-derived osteopontin reprograms normal mammary fibroblasts to promote inflammation and tumor growth in breast cancer. Cancer Res. 75, 963–973 (2015).
Serini, G. & Gabbiani, G. Mechanisms of myofibroblast activity and phenotypic modulation. Exp. Cell Res. 250, 273–283 (1999).
Flach, E. H., Rebecca, V. W., Herlyn, M., Smalley, K. S. & Anderson, A. R. Fibroblasts contribute to melanoma tumor growth and drug resistance. Mol. Pharm. 8, 2039–2049 (2011).
Gallagher, P. G. et al. Gene expression profiling reveals cross-talk between melanoma and fibroblasts: implications for host-tumor interactions in metastasis. Cancer Res. 65, 4134–4146 (2005).
Öhlund, D., Elyada, E. & Tuveson, D. Fibroblast heterogeneity in the cancer wound. J. Exp. Med. 211, 1503–1523 (2014).
Mitra, A. K. et al. MicroRNAs reprogram normal fibroblasts into cancer-associated fibroblasts in ovarian cancer. Cancer Discov. 2, 1100–1108 (2012).
Ruiter, D., Bogenrieder, T., Elder, D. & Herlyn, M. Melanoma–stroma interactions: structural and functional aspects. Lancet Oncol. 3, 35–43 (2002).
Haass, N. K., Smalley, K. S., Li, L. & Herlyn, M. Adhesion, migration and communication in melanocytes and melanoma. Pigment Cell Res. 18, 150–159 (2005).
Sugimoto, H., Mundel, T. M., Kieran, M. W. & Kalluri, R. Identification of fibroblast heterogeneity in the tumor microenvironment. Cancer Biol. Ther. 5, 1640–1646 (2006).
Hoashi, T. et al. Glycoprotein nonmetastatic melanoma protein b, a melanocytic cell marker, is a melanosome-specific and proteolytically released protein. FASEB J. 24, 1616–1629 (2010).
Torisu, H. et al. Macrophage infiltration correlates with tumor stage and angiogenesis in human malignant melanoma: possible involvement of TNFα and IL-1α. Int. J. Cancer 85, 182–188 (2000).
Mathivanan, S. & Simpson, R. J. ExoCarta: a compendium of exosomal proteins and RNA. Proteomics 9, 4997–5000 (2009).
Sato, I., Miyado, M. & Sunohara, M. NADH dehydrogenase activity and expression of mRNA of complex I (ND1, 51kDa, and 75kDa) in heart mitochondria of klotho mouse. Okajimas Folia Anat. Jpn. 82, 49–56 (2005).
Barker, H. E., Bird, D., Lang, G. & Erler, J. T. Tumor-secreted LOXL2 activates fibroblasts through FAK signaling. Mol. Cancer Res. 11, 1425–1436 (2013).
Bellei, B., Pitisci, A., Migliano, E., Cardinali, G. & Picardo, M. Pyridinyl imidazole compounds interfere with melanosomes sorting through the inhibition of cyclin G-associated Kinase, a regulator of cathepsins maturation. Cell. Signal. 26, 716–723 (2014).
Peinado, H. et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat. Med. 18, 883–891 (2012).
Xiao, D. et al. Identifying mRNA, microRNA and protein profiles of melanoma exosomes. PLoS ONE 7, e46874 (2012).
Lu, Y. J. et al. A molecular fluorescent dye for specific staining and imaging of RNA in live cells: a novel ligand integration from classical thiazole orange and styryl compounds. Chem. Commun. (Camb) 51, 15241–15244 (2015).
Sun, V., Zhou, W., Majid, S., Kashani-Sabet, M. & Dar, A. MicroRNA-mediated regulation of melanoma. Br. J. Dermatol. 171, 234–241 (2014).
Leibowitz-Amit, R., Sidi, Y. & Avni, D. Aberrations in the micro-RNA biogenesis machinery and the emerging roles of micro-RNAs in the pathogenesis of cutaneous malignant melanoma. Pigment Cell Melanoma Res. 25, 740–757 (2012).
Zhao, F. et al. Activation of p38 mitogen-activated protein kinase drives dendritic cells to become tolerogenic in ret transgenic mice spontaneously developing melanoma. Clin. Cancer Res. 15, 4382–4390 (2009).
Hankins, G. R. et al. M6P/IGF2 receptor: a candidate breast tumor suppressor gene. Oncogene 12, 2003–2009 (1996).
Oates, A. J. et al. The mannose 6-phosphate/insulin-like growth factor 2 receptor (M6P/IGF2R), a putative breast tumor suppressor gene. Breast Cancer Res. Treat. 47, 269–281 (1998).
Chappell, S., Walsh, T., Walker, R. & Shaw, J. Loss of heterozygosity at the mannose 6-phosphate insulin-like growth factor 2 receptor gene correlates with poor differentiation in early breast carcinomas. Br. J. Cancer 76, 1558 (1997).
Gemma, A. et al. Mutation analysis of the gene encoding the human mannose 6-phosphate/insulin-like growth factor 2 receptor (M6P/IGF2R) in human cell lines resistant to growth inhibition by transforming growth factor β 1 (TGF-β 1). Lung Cancer 30, 91–98 (2000).
Aprelikova, O. et al. The role of miR-31 and its target gene SATB2 in cancer-associated fibroblasts. Cell Cycle 9, 4387–4398 (2010).
Golan, T. et al. Interactions of melanoma cells with distal keratinocytes trigger metastasis via notch signaling inhibition of MITF. Mol. Cell (2015).
Bell, R. E. et al. Transcription factor/microRNA axis blocks melanoma invasion program by miR-211 targeting NUAK1. J. Invest. Dermatol. 134, 441–451 (2014)10.1038/jid.2013.340.
Soderberg, A., Barral, A. M., Soderstrom, M., Sander, B. & Rosen, A. Redox-signaling transmitted in trans to neighboring cells by melanoma-derived TNF-containing exosomes. Free Radic. Biol. Med. 43, 90–99 (2007).
Gu, J. et al. Gastric cancer exosomes trigger differentiation of umbilical cord derived mesenchymal stem cells to carcinoma-associated fibroblasts through TGF-β/Smad pathway. PLoS ONE 7, e52465 (2012).
van den Boorn, J. G. et al. Skin-depigmenting agent monobenzone induces potent T-cell autoimmunity toward pigmented cells by tyrosinase haptenation and melanosome autophagy. J. Invest. Dermatol. 131, 1240–1251 (2011).
Pinner, S. et al. Intravital imaging reveals transient changes in pigment production and Brn2 expression during metastatic melanoma dissemination. Cancer Res. 69, 7969–7977 (2009).
Lazar, I. et al. Proteome characterization of melanoma exosomes reveals a specific signature for metastatic cell lines. Pigment Cell Melanoma Res. 28, 464–475 (2015).
D’Souza-Schorey, C. & Clancy, J. W. Tumor-derived microvesicles: shedding light on novel microenvironment modulators and prospective cancer biomarkers. Genes Dev. 26, 1287–1299 (2012).
Morris, J. K. A formaldehyde glutaraldehyde fixative of high osmolality for use in electron microscopy. J. Cell Biol. 27, 137–139 (1965).
Ros, J. R., Rodriguez-Lopez, J. N. & Garcia-Canovas, F. Effect of L-ascorbic acid on the monophenolase activity of tyrosinase. Biochem. J. 295, 309–312 (1993).
Ritchie, M. E., Dunning, M. J., Smith, M. L., Shi, W. & Lynch, A. G. BeadArray expression analysis using bioconductor. PLoS Comput. Biol. 7, e1002276 (2011).
Kallio, M. A. et al. Chipster: user-friendly analysis software for microarray and other high-throughput data. BMC Genomics 12, 507 (2011).
Kramer, A., Green, J., Pollard, J. Jr & Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 30, 523–530 (2014).
Alhamdani, M. S. et al. Single-step procedure for the isolation of proteins at near-native conditions from mammalian tissue for proteomic analysis on antibody microarrays. J. Proteome Res. 9, 963–971 (2010).
Bogatkevich, G. S., Tourkina, E., Silver, R. M. & Ludwicka-Bradley, A. Thrombin differentiates normal lung fibroblasts to a myofibroblast phenotype via the proteolytically activated receptor-1 and a protein kinase C-dependent pathway. J. Biol. Chem. 276, 45184–45192 (2001).
Magina, S. et al. Inhibition of basal and ultraviolet B-induced melanogenesis by cannabinoid CB(1) receptors: a keratinocyte-dependent effect. Arch. Dermatol. Res. 303, 201–210 (2011).
Pons, M., Foradada, M. & Estelrich, J. Liposomes obtained by the ethanol injection method. Int. J. Pharm. 95, 51–56 (1993).
Acknowledgements
This work was supported in part by the Cooperation Program in Cancer Research of the Deutsches Krebsforschungszentrum (DKFZ) and Israel’s Ministry of Science, Technology and Space (MOST) and by the Israel Science Foundation (ISF). We thank the microarray unit of the DKFZ Genomics and Proteomics Core Facility for performing microarray analyses on the Illumina Human Sentrix-12 BeadChip array and the Agilent Human miRNA Microarray.
Author information
Authors and Affiliations
Contributions
S.D. designed the experimental approach, performed experimental work, analysed data and wrote the manuscript. L.S. conducted experimental work, miRNA and gene expression studies, exosomes analysis, and analysis of the microarray data. H.S. and N.E. conducted in vivo experiments, and FACS sorting, and added discussion on CAFs formation, D.S., Y.D. and H.M. conducted melanosome isolation and RNA detection experiments. A.Zilberberg conducted immunofluorescence staining and tyrosinase activity assays. L.L. helped in establishing the melanosome isolation protocol. S.G., A.B., I.B., R.B., P.G. and H.V. provided human skin and melanoma specimens and edited the manuscript. A.Zinger and A.S. generated the labelled liposomes and performed their analysis. T.G. performed the liposome uptake analysis. O.B. provided critical discussion and aided in writing of the manuscript. Y.F. and M.N. performed the electronic microscopy analysis. V.I. provided normal human melanocytes. J.D.H. coordinated the project, analysed and provided gene expression study data, and participated in writing of the manuscript. C.L. developed the hypothesis, designed the experimental approach, coordinated the project, and wrote the manuscript. S.A. conducted the exosome experiment analysis. M.K. designed and cloned the tyrosinase:mCherry plasmid.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
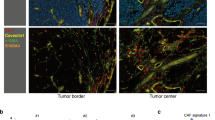
Supplementary Figure 1 Fibroblasts aggregation at a distance from melanoma regions contain melanoma particles.
(a–d) Immunofluorescence analysis of sections from four melanoma patients. DAPI-stained nuclei appear in blue. (e) Left: Immunohistochemistry staining (20X magnification) of MITF and MelanA in in situ melanoma sample. Right: Immunofluorescence analysis (63X magnification) of DCT (green) and HMB-45 (red) or FSP-1 (green) and HMB-45 (red). (f) Left: H&E staining (20X magnification) of in situ melanoma. Right: Immunofluorescence analysis (63X magnification) of HMB-45 (red) and DCT (green) or FSP-1 (green). Yellow signal indicates co-localization. (g) Left: H&E staining, immunohistochemistry for MITF and MelanA for in situ melanoma (20X magnification). Right: Immunofluorescence analysis of GPNMB (red) and FSP-1 (green). Yellow signal indicates co-localization of FSP-1 and GPNMB (63X magnification), in dashed square higher magnification image (100x magnification). (h) From left to right in in situ melanoma: H&E staining, immunohistochemistry (20X magnification) for MelanA, immunofluorescence analysis of HMB-45 (red) and DCT (green), HMB-45 (red) and FSP-1 (green), GPNMB (red) and FSP-1 (green). Yellow signal indicates co-localization (63X magnification).
Supplementary Figure 2 Fibroblasts aggregation at a distance from melanoma regions contain melanoma particles.
(a) Normal skin stained for HMB-45 (red) and DCT (green). DAPI-stained nuclei appear in blue (63X magnification). (b) Immunohistochemistry for MITF and MelanA for invasive melanoma (20X magnification) shown in Figure 1. (c) Immunofluorescence analysis of FSP-1 (green) and GPNMB (red) for invasive melanoma (20X magnification). Higher magnification image in the lower panel (100X magnification). (d) Left: H&E staining and immunohistochemistry for MelanA (20X magnification) for invasive melanoma. Right: Immunofluorescence analysis of DCT (green) and HMB-45 (red) for invasive melanoma (63X magnification). (e) Melanosome quantification method. Immunofluorescence analysis of dermal fibroblasts in superficial melanoma patient samples reveals staining for GPNMB (red) indicative of melanosomes. Left: Red fluorescence regions within fibroblast cytoplasm at the size of 300-600 nm are marked and were analyzed for fluorescence intensity (n = 80 melanosomes). Right: Whole cell fibroblasts were marked and analyzed for fluorescence intensity (n = 100 cells). (f) A single representative sample is shown stained for FSP-1 (green) and macrophage marker CD68 (red). White arrow indicates non-specific staining, and yellow signal indicates co-localization of FSP-1 and CD68. DAPI-stained nuclei appear in blue.
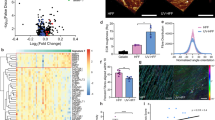
Supplementary Figure 3 Transfer of melanoma melanosomes into fibroblasts leads to reprogramming.
(a) Tyrosinase activity of MNT-1 whole cell extracts or isolated mature and pre-mature melanosomes. Values are normalized to total protein concentration. Error bars represent ± SEM, ∗ indicates p < 0.05 (n = 5 independent experiments). (b) Tyrosinase protein levels in indicated fractions. Rab27a was analyzed as a loading control. (c) Electron microscopy analysis of exosomes. (d) Expression of CD63 and CD81 in exosomes purified from MNT-1 cells by FACS analysis. Upper panel shows the analysis of expression of CD63 (blue) compared to unstained exosomes (red). Middle panel shows the analysis of expression of CD81 (blue) compared to unstained exosomes (red). Lower panel shows the analysis of expression of exosome markers CD63 and CD81 (blue) compared to unstained exosomes (red). (e) Protein levels in melanosomes isolated from MNT-1 cell homogenates and exosomes isolated from MNT-1 conditioned medium. Hsc70 was used as a loading control. (f) ND-1 levels in indicated fractions, normalized to GAPDH (n = 1 independent experiment). (g) Immunofluorescence staining of HaCat cells upon melanosome treatment. Melanosomal markers: tyrosinase, Tyrp1, and HMB-45 appear in yellow, red, and green, respectively. DAPI-stained nuclei appear blue. (h) Standard curves of melanosome number based on melanin and protein concentrations. (i) Fibroblasts migration upon osteopontin (400 nM) treatment. Unprocessed scans of Western Blots are shown in Supplementary Fig. 8.
Supplementary Figure 4 Melanoma melanosomes transfer into fibroblasts leads to reprogramming.
(a) Dynamic light scattering image of liposomes from a NanoSight analysis. Middle: Snapshot image of liposomes movie (Supplementary video 1). (b) Fibroblast migration upon indicated treatments. (c) Growth rate of fibroblasts upon indicated treatments (n = 2 independent experiments). (d,e) The pro-inflammatory gene signature of CAFs upon indicated treatments. Data were normalized to GAPDH (n = 3 independent experiments). (f) Left: Representative images of fibroblasts treated with 500 rhodamine-labeled liposomes or melanosomes (immunostained with GPNMB) per cell. Right: Average number of vesicles per cell (n = 6 cells for each group, 2 independent experiments). (g) Standard curve of liposome number. (h) Left: Number of liposomes per cell, calculated based on the standard curve (n = 3 measurements each of 9∗ 103 cells). Right: Representative image of fibroblasts treated with 500 rhodamine-labeled liposomes per cell. (i) Left: Electron microscopy analysis of secreted vesicles. Right: Higher magnification of the same sample. Red arrows indicate exosomes. Graphs: Percentages of indicated vesicles (n = 3 individual experiments). (j) Electron microscopy analysis of secreted melanosomes. (k) FACS analysis of CD63 and CD81 expression in indicated vesicles. Unstained MNT-1 cells appear in pale purple, stained MNT-1 cells in purple, unstained MNT-1 exosomes in pale red, stained MNT-1 exosomes in red, unstained MNT-1 melanosomes in pale blue, and stained MNT-1 melanosomes in blue. (l) NanoSight analysis of the secreted vesicle population upon P38 inhibitor treatment (n = 3 independent experiments). (m) Left: Melanin levels in melanoma medium upon SB202190 treatment. Data were normalized to RNU6B (n = 4 independent experiments). Right: Melanin levels in fibroblasts treated with medium from melanoma cells incubated with or without SB202190. Fibroblast medium was used as a control. Values were normalized to total protein amount (n = 8 samples, 3 independent experiments). (n) Immunostaining of fibroblasts treated with medium from melanoma cells that were pre-treated with SB202190. Tyrosinase is stained green; DAPI-stained nuclei appear in blue. (o) Bright field pictures of melanoma cells upon SB202190 treatment. (p) Growth rates of melanoma cells upon SB202190 treatment. (n = 3 independent experiments). (q,r) Migration and pro-inflammatory gene signature of fibroblasts treated with SB202190. Data were normalized to GAPDH (n = 2 independent experiments). In all relevant panels error bars represent ± SEM; ∗ indicates p < 0.05.
Supplementary Figure 5 Identification of melanosomal miRNAs and their trafficking into fibroblasts.
(a) Venn diagram showing the overlap between the 100 most highly expressed miRNAs in melanosomes, exosomes and melanoma-related miRNAs (literature based 41,42). (b) Yield of mature miRNAs in preparations from whole cells, mature melanosomes, and early melanosomes. Data were normalized to levels of RNU6B. Error bars represent ± SEM (n = 3 independent experiments). (c) Levels of indicated mature miRNAs were evaluated in fibroblasts treated with melanosomes or PBS (control) using RT-PCR. Data were normalized to RNU6B levels. Error bars represent ± SEM, ∗ indicates p < 0.05 (n = 4 independent experiments). (d) Table of the five microRNAs that are most up-regulated in mature melanosomes compared to their levels in whole cells. (e) Expression level of mature mmu-miR-211 in human fibroblasts after treatment with B16F10 melanosomes was determined using qRT-PCR. Data were normalized to RNU6B (n = 2 independent experiments). (f) Human primary fibroblasts were transfected with human or mouse miR-211 mimic (hsa and mmu, respectively). qRT- PCR analysis of both miRNAs. Data were normalized to RNU6B (n = 2 independent experiments). (g) Mature miRNAs levels in normal and melanoma melanosomes (n = 2 independent experiments). (h) Levels of mature miRNAs in indicated fractions (n = 2 independent experiments). (i) Levels of mature miRNAs in secreted melanosomes treated with the inhibitor (SB202190), normalized to RNU6B (n = 2 independent experiments).
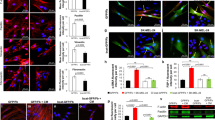
Supplementary Figure 6 Fibroblast reprogramming into CAFs is melanosomal miR-211 dependent.
Fibroblasts treated with miR-320c were analyzed in comparison to control fibroblasts treated with scrambled miRs. The fibroblasts were subjected to the following analyses: (a) growth rate (error bars represent ± SEM, n = 4 independent experiments); (b) migration tested by scratch assay; (c) fibroblast collagen remodeling capacity examined by three-dimensional collagen contraction assay (error bars represent ± SEM, n = 5 independent experiments); and (d) pro-inflammatory gene expression levels tested by qRT-PCR normalized to GAPDH levels (error bars represent ± SEM,∗ indicates p < 0.05, n = 3 independent experiments). (e) In situ hybridization analysis (green, 20x magnification) of miR-211 expression levels in healthy skin sample (left) and sections from the skin of a melanoma patient (right). Immunofluorescence staining of HMB-45 of the same magnification and location (red). DAPI-stained nuclei appear blue. (f) Mature mouse miR-211 levels (mmu-miR-211) were measured in human fibroblasts that were treated with medium of Ret melanoma cell line or with control medium. Data were normalized to RNU6B levels. Error bars represent ± SEM, ∗ indicates p < 0.05 (n = 3 independent experiments). (g) PDGFRα-positive fibroblasts from local tumor stroma and melanoma cells from tumor were assessed for the expression of fibroblast markers (α-SMA, collagen), macrophage markers (CD45, F40/80), and melanoma markers (trpm1, tyrosinase). Results were compared to the expression in normal fibroblasts sorted from mouse ears and normalized to GAPDH. Error bars represent ± SEM, ∗ indicates p < 0.05 (n = 20 mice for treatment and 12 for control in 3 independent experiments). (h) Sorted fibroblasts from normal and stromal tissue stained for tyrosinase (green). DAPI-stained nuclei appear blue. Left panel: 40x magnification, right panel: 100x magnification.
Supplementary Figure 7 Melanosomal miR-211 induces CAF formation via up-regulation of MAPK signaling.
(a) Hierarchical clustering analysis of mRNA expression profiles of fibroblasts upon indicated treatments. Each row represents the mean of signal log ratios using a color-code scale. (b) Canonical pathway for IGF1R signaling in fibroblasts upon indicated treatments, generated with IPA. Gene expression levels are relative to control fibroblasts. Up- and down-regulated genes are labeled red or green, respectively; predicted activation is shown in orange, inhibition in blue. Lines represent known interactions. Orange arrows: activation; blue arrows: inhibition; yellow arrows: inconsistency between observation and prediction; grey arrows: lack of supporting data. (c,d) Immunostaining of HMB-45 and IGF2R in human specimens. Dashed lines separate tumor (T) and stroma (S); epidermis (E) and dermis (D) are indicated. DAPI-stained nuclei appear blue.. (e) Left: Immunostaining of HMB-45 and IGF2R in human specimens. Right: In situ hybridization of miR-211. Dashed lines separate tumor (T) and stroma (S); epidermis (E) and dermis (D) are indicated. DAPI-stained nuclei appear blue (63X magnification). (f) Top: Immunostaining of HMB-45 and IGF2R in human specimens. Bottom: In situ hybridization of miR-211. Dashed lines separate tumor (T) and stroma (S); epidermis (E) and dermis (D) are indicated. DAPI-stained nuclei appear blue (63X magnification). (g) MNT-1 and WM3314 melanoma cells invasion abilities, measured with a Matrigel invasion assay. Graph plots percent of invading cells. Representative images of invading and seeded cells are shown; DAPI-stained nuclei appear in blue. Error bars represent ± SEM, ∗ indicates p < 0.05 (n = 3 independent experiments). (h) MITF expression levels in MNT-1 and WM3314 melanoma cells normalized to GAPDH. Error bars represent ± SEM (n = 3 independent experiments). (i) Venn diagram showing the overlap between the 100 most highly expressed miRNAs in MNT-1 and WM3314 mature melanosomes and melanoma-related miRNAs (literature based43,44). (j) Pearson’s correlation for the expression level of each miRNA relative to MITF in 11 melanoma cell lines. miR-211, Let7a, miR-17 and miR-125b are highlighted (correlations of p < 0.05). (k) A summary model - The formation of the melanoma primary niche by miRNA trafficking. Graphical model describing our findings which demonstrate that melanoma cells directly affect the formation of the dermal tumor niche by miRNA trafficking prior to invading the dermis.
Supplementary information
Supplementary Information
Supplementary Information (PDF 2871 kb)
Supplementary Table 1
Supplementary Information (XLSX 12245 kb)
Supplementary Table 2
Supplementary Information (XLSX 124 kb)
Supplementary Table 3
Supplementary Information (XLSX 1085 kb)
Supplementary Table 4
Supplementary Information (XLSX 220 kb)
Supplementary Table 5
Supplementary Information (XLSX 1236 kb)
Supplementary Table 6
Supplementary Information (XLSX 534 kb)
Supplementary Table 7
Supplementary Information (XLSX 13 kb)
Supplementary Table 8
Supplementary Information (XLSX 61 kb)
Supplementary Table 9
Supplementary Information (XLSX 11 kb)
Rights and permissions
About this article
Cite this article
Dror, S., Sander, L., Schwartz, H. et al. Melanoma miRNA trafficking controls tumour primary niche formation. Nat Cell Biol 18, 1006–1017 (2016). https://doi.org/10.1038/ncb3399
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb3399
This article is cited by
-
Define cancer-associated fibroblasts (CAFs) in the tumor microenvironment: new opportunities in cancer immunotherapy and advances in clinical trials
Molecular Cancer (2023)
-
Exosome application in tumorigenesis: diagnosis and treatment of melanoma
Medical Oncology (2022)
-
SEMA6A/RhoA/YAP axis mediates tumor-stroma interactions and prevents response to dual BRAF/MEK inhibition in BRAF-mutant melanoma
Journal of Experimental & Clinical Cancer Research (2022)
-
Signaling pathways in cancer-associated fibroblasts and targeted therapy for cancer
Signal Transduction and Targeted Therapy (2021)
-
Signal pathways of melanoma and targeted therapy
Signal Transduction and Targeted Therapy (2021)