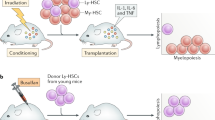
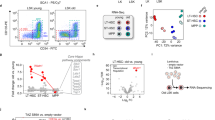
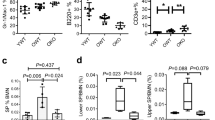
Abstract
Ageing-associated impairments in haemato-lymphopoiesis are associated with DNA damage accumulation and reduced maintenance of lymphoid-biased (Ly-biased) compared with myeloid-biased (My-biased) haematopoietic stem cells (HSCs). Here, in vivo RNAi screening identifies period circadian clock 2 (Per2) as a critical factor limiting the maintenance of HSCs in response to DNA damage and ageing. Under these conditions, Per2 is activated predominantly in Ly-biased HSCs and stimulates DNA damage signalling and p53-dependent apoptosis in haematopoietic cells. Per2 deletion ameliorates replication stress and DNA damage responses in haematopoietic cells, thereby improving the maintenance of Ly-biased HSCs, lymphopoiesis, and immune function in ageing mice without increasing the accumulation of DNA damage. Per2-deficient mice retain Batf/p53-dependent induction of differentiation of HSCs in response to DNA damage and exhibit an elongated lifespan. Together, these results identify Per2 as a negative regulator of Ly-biased HSCs and immune functions in response to DNA damage and ageing.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Rossi, D. J. et al. Cell intrinsic alterations underlie hematopoietic stem cell aging. Proc. Natl Acad. Sci. USA 28, 9194–9199 (2005).
Geiger, H., Rennebeck, G. & Van Zant, G. Regulation of hematopoietic stem cell aging in vivo by a distinct genetic element. Proc. Natl Acad. Sci. USA 102, 5102–5107 (2005).
Morrison, S. J., Wandycz, A. M., Akashi, K., Globerson, A. & Weissman, I. L. The aging of hematopoietic stem cells. Nat. Med. 2, 1011–1016 (1996).
Sudo, K., Ema, H., Morita, Y. & Nakauchi, H. Age-associated characteristics of murine hematopoietic stem cells. J. Exp. Med. 192, 1273–1280 (2000).
Wang, J. W., Geiger, H. & Rudolph, K. L. Immunoaging induced by hematopoietic stem cell aging. Curr. Opin. Immunol. 23, 532–536 (2011).
Gazit, R., Weissman, I. L. & Rossi, D. J. Hematopoietic stem cells and the aging hematopoietic system. Semin. Hematol. 45, 218–224 (2008).
Linton, P. J. & Dorshkind, K. Age-related changes in lymphocyte development and function. Nat. Immunol. 5, 133–139 (2004).
Cho, R. H., Sieburg, H. B. & Muller-Sieburg, C. E. A new mechanism for the aging of hematopoietic stem cells: aging changes the clonal composition of the stem cell compartment but not individual stem cells. Blood 111, 5553–5561 (2008).
Beerman, I. et al. Functionally distinct hematopoietic stem cells modulate hematopoietic lineage potential during aging by a mechanism of clonal expansion. Proc. Natl Acad. Sci. USA 107, 5465–5470 (2010).
Pang, W. W. et al. Human bone marrow hematopoietic stem cells are increased in frequency and myeloid-biased with age. Proc. Natl Acad. Sci. USA 108, 20012–20017 (2011).
Rossi, D. J. et al. Deficiencies in DNA damage repair limit the function of haematopoietic stem cells with age. Nature 447, 725–729 (2007).
Rübe, C. E. et al. Accumulation of DNA damage in hematopoietic stem and progenitor cells during human aging. PLoS ONE 6, e17487 (2011).
Ito, K. et al. Reactive oxygen species act through p38 MAPK to limit the lifespan of hematopoietic stem cells. Nat. Med. 12, 446–451 (2006).
Ito, K. et al. Regulation of oxidative stress by ATM is required for self-renewal of haematopoietic stem cells. Nature 431, 997–1002 (2004).
Flach, J. et al. Replication stress is a potent driver of functional decline in ageing haematopoietic stem cells. Nature 512, 198–202 (2014).
Walter, D. et al. Exit from dormancy provokes DNA-damage-induced attrition in haematopoietic stem cells. Nature 520, 549–552 (2015).
Choudhury, A. R. et al. Cdkn1a deletion improves stem cell function and lifespan of mice with dysfunctional telomeres without accelerating cancer formation. Nat. Genet. 39, 99–105 (2007).
Wang, J. W. et al. A differentiation checkpoint limits hematopoietic stem cell self-renewal in response to DNA damage. Cell 148, 1001–1014 (2012).
Vaziri, H. et al. Evidence for a mitotic clock in human hematopoietic stem cells: loss of telomeric DNA with age. Proc. Natl Acad. Sci. USA 91, 9857–9860 (1994).
Calado, R. T. & Young, N. S. Telomere maintenance and human bone marrow failure. Blood 111, 4446–4455 (2008).
Ohyashiki, J. H. et al. Telomere shortening associated with disease evolution patterns in myelodysplastic syndromes. Cancer Res. 54, 3557–3560 (1994).
Blasco, M. A. et al. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell 91, 25–34 (1997).
Rudolph, K. L. et al. Longevity, stress response, and cancer in aging telomerase-deficient mice. Cell 96, 701–712 (1999).
Herrera, E. et al. Disease states associated with telomerase deficiency appear earlier in mice with short telomeres. EMBO J. 18, 2950–2960 (1999).
Chin, L. et al. p53 deficiency rescues the adverse effects of telomere loss and cooperates with telomere dysfunction to accelerate carcinogenesis. Cell 97, 527–538 (1999).
Fu, L. N. et al. The Circadian Gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell 111, 41–50 (2002).
Beerman, I. et al. Proliferation-dependent alterations of the DNA methylation landscape underlie hematopoietic stem cell aging. Cell Stem Cell 12, 413–425 (2013).
Beerman, I., Seita, J., Inlay, M. A., Weissman, I. L. & Rossi, D. J. Quiescent hematopoietic stem cells accumulate DNA damage during aging that is repaired upon entry into cell cycle. Cell Stem Cell 15, 37–50 (2014).
Lee, Z. & Stephen, J. E. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300, 1542–1548 (2003).
Shao, R. G. et al. DNA damage by camptothecin induces phosphorylation of RPA by DNA-dependent protein kinase and dissociates RPA:DNA-PK complexes. EMBO J. 18, 1397–1406 (1999).
Vassin, V. M., Anantha, R. W., Sokolova, E., Kanner, S. & Borowiec, J. A. Human RPA phosphorylation by ATR stimulates DNA synthesis and prevents ssDNA accumulation during DNA-replication stress. J. Cell Sci. 122, 4070–4080 (2009).
Geiger, H. & Rudolph, K. L. Aging in the lympho-hematopoietic stem cell compartment. Trends Immunol. 30, 360–365 (2009).
Ju, Z. et al. Telomere dysfunction induces environmental alterations limiting hematopoietic stem cell function and engraftment. Nat. Med. 13, 742–747 (2007).
Song, Z. et al. Alterations of the systemic environment are the primary cause of impaired B and T lymphopoiesis in telomere-dysfunctional mice. Blood 115, 1481–1489 (2010).
Miller, J. P. & Allman, D. The decline in B lymphopoiesis in aged mice reflects loss of very early B-lineage precursors. J. Immunol. 171, 2326–2330 (2003).
Minegishi, Y. et al. An essential role for BLNK in human B cell development. Science 286, 1954–1957 (1999).
Setoguchi, R. et al. Homeostatic maintenance of natural Foxp3(+) CD25(+) CD4(+) regulatory T cells by interleukin (IL)-2 and induction of autoimmune disease by IL-2 neutralization. J. Exp. Med. 201, 723–735 (2005).
Hori, S. et al. Control of regulatory T cell development by the transcription factor Foxp3. Science 299, 1057–1061 (2003).
Kaufmann, C. et al. A complex network of regulatory elements in Ikaros and their activity during hemo-lymphopoiesis. EMBO J. 22, 2211–2223 (2003).
Kopf, M. et al. Impaired immune and acute-phase responses in interleukin-6-deficient mice. Nature 368, 339–342 (1994).
Puel, A., Ziegler, S. F., Buckley, R. H. & Leonard, W. J. Defective IL7R expression in T(-)B(+)NK(+) severe combined immunodeficiency. Nat. Genet. 20, 394–397 (1998).
Nurieva, R. et al. Essential autocrine regulation by IL-21 in the generation of inflammatory T cells. Nature 448, 480–483 (2007).
Simard, N. et al. Analysis of the role of IL-21 in development of murine B cell progenitors in the bone marrow. J. Immunol. 186, 5244–5253 (2011).
Limnander, A. et al. STIM1, PKC-δ and RasGRP set a threshold for proapoptotic Erk signaling during B cell development. Nat. Immunol. 12, 425–433 (2011).
Pasqualucci, L. et al. Inactivation of the PRDM1/BLIMP1 gene in diffuse large B cell lymphoma. J. Exp. Med. 203, 311–317 (2006).
Alsadeq, A., Hobeika, E., Medgyesi, D., Kläsener, K. & Reth, M. The role of the Syk/Shp-1 kinase-phosphatase equilibrium in B cell development and signaling. J. Immunol. 193, 268–276 (2014).
Veldhoen, M., Hocking, R. J., Atkins, C. J., Locksley, R. M. & Stockinger, B. TGFβ in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity 24, 179–189 (2006).
Hayashi, E. A. et al. Role of TLR in B cell development: signaling through TLR4 promotes B cell maturation and is inhibited by TLR2. J. Immunol. 174, 6639–6647 (2005).
Frasca, D., Diep, N. D., Riley, R. L. & Blomberg, B. B. Decreased E12/E47 transcription factor activity in the bone marrow as well as in spleen of aged mice. J. Immunol. 170, 2719–2726 (2003).
Vidarsson, G., Dekkers, G. & Rispens, T. IgG subclasses and allotypes: from structure to effector functions. Front Immunol. 5, 520 (2014).
Nippe, N. et al. Subcutaneous infection with S. aureus in mice reveals association of resistance with influx of neutrophils and Th2 response. J. Invest. Dermatol. 131, 125–132 (2011).
Donehower, L. A. et al. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature 356, 215–221 (1992).
Jacks, T. et al. Tumor spectrum analysis in p53-mutant mice. Curr. Biol. 4, 1–7 (1994).
Lee, S., Donehower, L. A., Herron, A. J., Moore, D. D. & Fu, L. Disrupting circadian homeostasis of sympathetic signaling promotes tumor development in mice. PLoS ONE 5, e10995 (2010).
Santos, M. A. et al. DNA-damage-induced differentiation of leukaemic cells as an anti-cancer barrier. Nature 514, 107–111 (2014).
Sangoram, A. M. et al. Mammalian circadian autoregulatory loop: a timeless ortholog and mPer1 interact and negatively regulate CLOCK-BMAL1-induced transcription. Neuron 21, 1101–1113 (1998).
Toh, K. L. et al. An hPer2 phosphorylation site mutation in familial advanced sleep-phase syndrome. Science 291, 1040–1043 (2001).
Ema, H. et al. Adult mouse hematopoietic stem cells: purification and single-cell assays. Nat. Protoc. 1, 2979–2987 (2007).
Acknowledgements
This work was supported by the European Union (advanced ERC grant to K.L.R., grant 323136—StemCellGerontoGenes), within the e:Med program (HaematoOpt) of the German Federal Ministry of Education and Research (BMBF), the German Research Foundation (DFG—RU745-10), the Baden-Württemberg-Stiftung (P1301029), the Leibniz association, and the State of Thuringia (FZ-12001-514). We thank Z.-Q. Wang from the Leibniz Institute on Aging for supplying ATM-deficient mice.
Author information
Authors and Affiliations
Contributions
J.W. performed most of the experiments, Y.M. performed FACS analysis and wrote the manuscript; B.H. performed ELISA measurements, B.L. and S.N. performed the food pad assay and K.L.R. designed and supervised the project, and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Verification of shRNAs of candidate genes.
(A) The representative Western blot shows the efficiency of knockdown (by shRNAs) and over-expression (O/E) of Per2. Cell lysates obtained from lineage bone marrow cells that were infected by lentiviral vectors carrying two different shRNA (shRNA1 or shRNA2) targeting Per2 or Per2-cDNA or an empty control vector were loaded to detect PER2 protein expression. Data represent 1 out 3 independent experiments. (B) The histogram shows the mRNA level of Pcdh20 in lineage bone marrow cells of WT mice infected by lentiviruses carrying two different shRNAs against Pcdh20 or a scramble shRNA control vector. N = 3 repeat experiments, values are means ± s.e.m. (C) The histogram depicts the number of shRNAs detected by deep sequencing in the shRNA plasmid pool as well as in Lin cells derived from mTerc+/+ and G3 mTerc−/− donors after two round of transplantation as depicted in Fig. 1a. (D,E) Freshly isolated KSL cells from G3 mTerc−/− mice or mTerc+/+ mice were infected with 2 independent single shRNA constructs targeting (D) Per2 or (E) Pcdh20, or a scrambled shRNA control. Infected cells were transplanted into lethally irradiated mice along with non-infected cells from the same culture. The graphs show the changes in the percentage of infected cells (GFP+) in peripheral blood of primary recipients at indicated time points after transplantation. N = 5 recipients/group, values are means ± s.e.m., multiple t test was used to calculate P values.
Supplementary Figure 2 Per2 deletion improves self-renewal of HSCs in response to replicative stress.
(A–C) Cohorts of 3-month-old Per2−/− mice and Per2+/+ mice were sub-lethally irradiated (4 Gy) to induce premature aging and kept for 1 year. At this time point mice were sacrificed to analyse hematopoietic cells in bone marrow (8 Per2+/+ mice and 7 Per2−/− mice). (A,B) The histograms show the absolute number of (A) Ly-biased HSCs (marked by CD150lo CD34 KSL) and (B) My-biased HSCs (marked by CD150hi CD34 KSL), values are shown as mean ± s.e.m., multiple t test was used to calculate P values. (C) Representative FACS plots showing an increase in Ly-biased HSCs (CD150lo, CD34) in bone marrow of Per2−/− mice versus Per2+/+ mice. FACS was repeated on biological replicates (8 times for Per2+/+ mice and 7 times for Per2−/− mice).
Supplementary Figure 3 Per2 contributes to the activation of apoptosis signalling in hematopoietic cells in response to DNA damage but not to DNA damage sensing.
(A) Immunofluorescence staining of γH2AX in freshly isolated HSCs (CD34 LSK) from irradiated (4 Gy, 6 h after IR) and non-irradiated (nIR), 2–3-month-old Per2+/+ mice or Per2−/− mice. Representative data derived from 200 analysed cells for 1 out of 5 mice per genotype are shown. Chi-square test was used to generate P values. Note, there is no impact of Per2 gene status on the induction of γH2AX foci. (B) 8000 freshly isolated KSL cells from 2-month-old Per2+/+ and Per2−/− mice were cultured for 12 h followed by sub-lethal irradiation (4 Gy). The numbers of living cells were counted 24 h and 36 h after IR by automatic cell counter. This histogram depicts the number of living hematopoietic stem and progenitor cells (KSL) at indicated time points after sub-lethal irradiation (4 Gy). N = 5 repeat experiments per genotype, values are shown as mean ± s.e.m. and multiple t test was used to calculate P value.
Supplementary Figure 4 Per2 deletion rescues myeloid/lymphoid skewing in the context of DNA damage.
(A–C) 3-month-old Per2−/− and Per2+/+ mice were exposed to 4 Gy γ-irradiation (IR) or non-irradiated (nIR) and were analysed one year later (same as in Supplementary Fig. 2). (8 Per2+/+ mice and 7 Per2−/− mice per group), values are shown as mean ± s.e.m. Multiple t test was used to calculate P value. (A) This histogram shows the percentage of B220+ and CD11b+ cells in peripheral blood. (B) This histogram shows the percentage of B220+ and CD11b+ cells in bone marrow. (C) Representative FACS plots showing the B220+ and CD11b+ cells in bone marrow of mice of the indicated genotype and treatment.
Supplementary Figure 5 Per2 deletion does not change B-lymphocyte production of stimulated B cell progenitors from young mice.
(A) 1000 freshly purified EBPs from 2-month-old Per2−/− and Per2+/+ mice were cultured and exposed to IL-7 at the indicated concentrations. FACS analysis determined the total numbers of B220+CD19+ cells after 4-day culture. N = 5 mice per group, dots represent individual mice, lines depict mean values. In contrast to the results on EBPs from old mice (Fig. 5b), Per2 gene status does not affect B-lymphocyte production rates of stimulated B-cell progenitors from young mice.
Supplementary information
Supplementary Information
Supplementary Information (PDF 905 kb)
Supplementary Table 1
Supplementary Information (XLS 805 kb)
Supplementary Table 2
Supplementary Information (XLSX 9 kb)
Rights and permissions
About this article
Cite this article
Wang, J., Morita, Y., Han, B. et al. Per2 induction limits lymphoid-biased haematopoietic stem cells and lymphopoiesis in the context of DNA damage and ageing. Nat Cell Biol 18, 480–490 (2016). https://doi.org/10.1038/ncb3342
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb3342
This article is cited by
-
PER2: a potential molecular marker for hematological malignancies
Molecular Biology Reports (2021)
-
Elevated Hedgehog activity contributes to attenuated DNA damage responses in aged hematopoietic cells
Leukemia (2020)
-
Circadian regulation of mitochondrial uncoupling and lifespan
Nature Communications (2020)
-
Time after time: circadian clock regulation of intestinal stem cells
Cellular and Molecular Life Sciences (2020)
-
Hierarchically related lineage-restricted fates of multipotent haematopoietic stem cells
Nature (2018)