Abstract
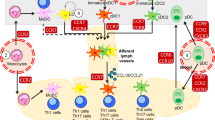
Dendritic cell (DC) migration in peripheral tissues serves two main functions: antigen sampling by immature DCs, and chemokine-guided migration towards lymphatic vessels (LVs) on maturation. These migratory events determine the efficiency of the adaptive immune response. Their regulation by the core cell locomotion machinery has not been determined. Here, we show that the migration of immature DCs depends on two main actin pools: a RhoA–mDia1-dependent actin pool located at their rear, which facilitates forward locomotion; and a Cdc42–Arp2/3-dependent actin pool present at their front, which limits migration but promotes antigen capture. Following TLR4–MyD88-induced maturation, Arp2/3-dependent actin enrichment at the cell front is markedly reduced. Consequently, mature DCs switch to a faster and more persistent mDia1-dependent locomotion mode that facilitates chemotactic migration to LVs and lymph nodes. Thus, the differential use of actin-nucleating machineries optimizes the migration of immature and mature DCs according to their specific function.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Change history
23 December 2015
In the version of this Article originally published, references 42 and 43 were incorrect. They have been replaced with a new ref. 42, and references 44–51 have been renumbered accordingly. These changes have been made in all online versions of the Article.
References
Mellman, I. & Steinman, R. M. Dendritic cells: specialized and regulated antigen processing machines. Cell 106, 255–258 (2001).
Garrett, W. S. et al. Developmental control of endocytosis in dendritic cells by Cdc42. Cell 102, 325–334 (2000).
Cella, M., Sallusto, F. & Lanzavecchia, A. Origin, maturation and antigen presenting function of dendritic cells. Curr. Opin. Immunol. 9, 10–16 (1997).
Sallusto, F., Cella, M., Danieli, C. & Lanzavecchia, A. Dendritic cells use macropinocytosis and the mannose receptor to concentrate macromolecules in the major histocompatibility complex class II compartment: downregulation by cytokines and bacterial products. J. Exp. Med. 182, 389–400 (1995).
Farache, J. et al. Luminal bacteria recruit CD103 + dendritic cells into the intestinal epithelium to sample bacterial antigens for presentation. Immunity 38, 581–595 (2013).
Ng, L. G. et al. Migratory dermal dendritic cells act as rapid sensors of protozoan parasites. PLoS Pathog. 4, e1000222 (2008).
Turley, S. J. et al. Transport of peptide-MHC class II complexes in developing dendritic cells. Science 288, 522–527 (2000).
MartIn-Fontecha, A. et al. Regulation of dendritic cell migration to the draining lymph node: impact on T lymphocyte traffic and priming. J. Exp. Med. 198, 615–621 (2003).
Tal, O. et al. DC mobilization from the skin requires docking to immobilized CCL21 on lymphatic endothelium and intralymphatic crawling. J. Exp. Med. 208, 2141–2153 (2011).
Weber, M. et al. Interstitial dendritic cell guidance by haptotactic chemokine gradients. Science 339, 328–332 (2013).
Acton, S. E. et al. Podoplanin-rich stromal networks induce dendritic cell motility via activation of the C-type lectin receptor CLEC-2. Immunity 37, 276–289 (2012).
Pollard, T. D. & Borisy, G. G. Cellular motility driven by assembly and disassembly of actin filaments. Cell 112, 453–465 (2003).
Lammermann, T. et al. Rapid leukocyte migration by integrin-independent flowing and squeezing. Nature 453, 51–55 (2008).
Renkawitz, J. et al. Adaptive force transmission in amoeboid cell migration. Nat. Cell Biol. 11, 1438–1443 (2009).
Faure-André, G. et al. Regulation of dendritic cell migration by CD74, the MHC class II-associated invariant chain. Science 322, 1705–1710 (2008).
Heuzé, M. L. et al. Migration of dendritic cells: physical principles, molecular mechanisms, and functional implications. Immunol. Rev. 256, 240–254 (2013).
Vargas, P., Terriac, E., Lennon-Dumenil, A. M. & Piel, M. Study of cell migration in microfabricated channels. J. Vis. Exp. 84, e51099 (2014).
Barton, G. M. & Medzhitov, R. Toll-like receptor signaling pathways. Science 300, 1524–1525 (2003).
Heit, B. & Kubes, P. Measuring chemotaxis and chemokinesis: the under-agarose cell migration assay. Sci. STKE 2003, PL5 (2003).
Chabaud, M. et al. Cell migration and antigen capture are antagonistic processes coupled by myosin II in dendritic cells. Nat. Commun. 6, 7526 (2015).
Bailly, M. et al. The F-actin side binding activity of the Arp2/3 complex is essential for actin nucleation and lamellipod extension. Curr. Biol. 11, 620–625 (2001).
Blanchoin, L. et al. Direct observation of dendritic actin filament networks nucleated by Arp2/3 complex and WASP/Scar proteins. Nature 404, 1007–1011 (2000).
Svitkina, T. M. & Borisy, G. G. Arp2/3 complex and actin depolymerizing factor/cofilin in dendritic organization and treadmilling of actin filament array in lamellipodia. J. Cell Biol. 145, 1009–1026 (1999).
Koivusalo, M. et al. Amiloride inhibits macropinocytosis by lowering submembranous pH and preventing Rac1 and Cdc42 signaling. J. Cell Biol. 188, 547–563 (2010).
Nolen, B. J. et al. Characterization of two classes of small molecule inhibitors of Arp2/3 complex. Nature 460, 1031–1034 (2009).
Goley, E. D. et al. An actin-filament-binding interface on the Arp2/3 complex is critical for nucleation and branch stability. Proc. Natl Acad. Sci. USA 107, 8159–8164 (2010).
Higashida, C. et al. Actin polymerization-driven molecular movement of mDia1 in living cells. Science 303, 2007–2010 (2004).
Li, F. & Higgs, H. N. The mouse Formin mDia1 is a potent actin nucleation factor regulated by autoinhibition. Curr. Biol. 13, 1335–1340 (2003).
Pruyne, D. et al. Role of formins in actin assembly: nucleation and barbed-end association. Science 297, 612–615 (2002).
Rizvi, S. A. et al. Identification and characterization of a small molecule inhibitor of formin-mediated actin assembly. Chem. Biol. 16, 1158–1168 (2009).
Eisenmann, K. M. et al. T cell responses in mammalian diaphanous-related formin mDia1 knock-out mice. J. Biol. Chem. 282, 25152–25158 (2007).
Sakata, D. et al. Impaired T lymphocyte trafficking in mice deficient in an actin-nucleating protein, mDia1. J. Exp. Med. 204, 2031–2038 (2007).
Tanizaki, H. et al. Rho-mDia1 pathway is required for adhesion, migration, and T-cell stimulation in dendritic cells. Blood 116, 5875–5884 (2010).
Bovellan, M. et al. Cellular control of cortical actin nucleation. Curr. Biol. 24, 1628–1635 (2014).
Peng, J., Wallar, B. J., Flanders, A., Swiatek, P. J. & Alberts, A. S. Disruption of the Diaphanous-related formin Drf1 gene encoding mDia1 reveals a role for Drf3 as an effector for Cdc42. Curr. Biol. 13, 534–545 (2003).
Machesky, L. M. et al. Scar, a WASp-related protein, activates nucleation of actin filaments by the Arp2/3 complex. Proc. Natl Acad. Sci. USA 96, 3739–3744 (1999).
Luckashenak, N., Wahe, A., Breit, K., Brakebusch, C. & Brocker, T. Rho-family GTPase Cdc42 controls migration of Langerhans cells in vivo. J. Immunol. 190, 27–35 (2013).
Sarris, M. et al. Inflammatory chemokines direct and restrict leukocyte migration within live tissues as glycan-bound gradients. Curr. Biol. 22, 2375–2382 (2012).
Weber, M. & Sixt, M. Live cell imaging of chemotactic dendritic cell migration in explanted mouse ear preparations. Methods Mol. Biol. 1013, 215–226 (2013).
Hawkins, R. J. et al. Pushing off the walls: a mechanism of cell motility in confinement. Phys. Rev. Lett. 102, 058103 (2009).
Wilson, K. et al. Mechanisms of leading edge protrusion in interstitial migration. Nat. Commun. 4, 2896 (2013).
Veltman, D. M. et al. PIP3-dependent macropinocytosis is incompatible with chemotaxis. J. Cell Biol. 204, 497–505 (2014).
Forster, R. et al. CCR7 coordinates the primary immune response by establishing functional microenvironments in secondary lymphoid organs. Cell 99, 23–33 (1999).
Riedl, J. et al. Lifeact mice for studying F-actin dynamics. Nat. Methods 7, 168–169 (2010).
Muzumdar, M. D., Tasic, B., Miyamichi, K., Li, L. & Luo, L. A global double-fluorescent Cre reporter mouse. Genesis 45, 593–605 (2007).
Wolf, K. et al. Physical limits of cell migration: control by ECM space and nuclear deformation and tuning by proteolysis and traction force. J. Cell Biol. 201, 1069–1084 (2013).
Preibisch, S., Saalfeld, S. & Tomancak, P. Globally optimal stitching of tiled 3D microscopic image acquisitions. Bioinformatics 25, 1463–1465 (2009).
Maiuri, P. et al. The first world cell race. Curr. Biol. 22, R673–R675 (2012).
R-Core-Team. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2014); http://www.r-project.org
Selmeczi, D., Mosler, S., Hagedorn, P. H., Larsen, N. B. & Flyvbjerg, H. Cell motility as persistent random motion: theories from experiments. Biophys. J. 89, 912–931 (2005).
Acknowledgements
The authors thank the PICT IBiSA platform at Institut Curie (CNRS UMR144, especially V. Fraisier) and the Institut Curie animal facility. P.V. thanks B. Goic for support, patience and critical reading of the manuscript. P.V. was supported by fellowships from Region Ile-de-France, Fondation pour la Recherche Médicale (FRM) and Institut Curie. M.B. and M.C. benefited from fellowships from Association pour la Recherche contre le Cancer and FRM respectively. S.M.K.-G. and A.S.A. were supported by the Grand Rapids Community and the Lunn Hope Foundations, the Van Andel Endowment, and the Purple Community. T.B. was supported by DFG SFB 1054-B03 and SFB 914-A06. This work was financially supported by grants from: the City of Paris, the European Research Council and the DCBIOL Labex (ANR-10-IDEX-0001-02-PSL∗ and ANR-11-LABX-0043) to A.-M.L.-D. (Strapacemi 243103) and the Association Nationale pour la Recherche (ANR-09-PIRI-0027-PCVI) and the InnaBiosanté foundation (Micemico) to A.-M.L.-D., M.P. and R.V.
Author information
Authors and Affiliations
Contributions
P.V. designed, performed and analysed most experiments, prepared all manuscript figures and strongly participated in article writing and project conception. P.M. conceived and performed trajectory analyses and modelling. M.B. set up and assisted with siRNA-mediated silencing, qPCR and macropinocytosis experiments. P.J.S. and H.-R.T. assisted in optimizing micro-channel experiments. P.J.S. further helped P.V. with article revision. P.P. built the program for actin distribution analysis in migrating DCs. M.M. created all codes for quantification of macropinocytosis and arrival of DCs to LVs. M.C. carried out the initial observation of increased migration speed and decreased antigen uptake in CK666-treated iDCs. E.T. helped with micro-fabrication. D.L. assisted P.V. with immunofluorescence experiments. D.O. performed immunoblots. M.R. set up collagen gel experiments. T.B. generated and provided Cdc42 and RhoA conditional KO mice. S.M.K.-G. and A.S.A. generated and provided mDia1KO mice. S.X., P.S. and R.L. generated and provided Arpc2 conditional KO mice. R.V. conceived and performed migration modelling. M.P. and A.-M.L.-D. designed the overall research and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 DC migration becomes faster and more persistent upon LPS treatment.
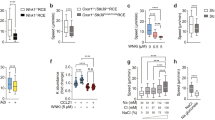
(a,b) Analysis by flow cytometry of surface expression of the DC activation marker CD86 and the chemokine receptor CCR7. iDCs were incubated for different times with LPS (100 ng/ml), washed and cultured overnight. 30 min was found to be the minimal time to get the highest level of activation and CCR7 expression. 1 representative experiment out of 3 is shown. (c) Mean instantaneous speed of migrating DCs in the presence of Pertussis toxin (Ptx, 200 μg/ml) in fibronectin- (c) or fibronectin plus CCL21- (d) coated micro-channels (n = 80, 94 and 91 for iDC, LPS-DC and LPS-DC Ptx respectively). 1 representative experiment out of 2 is shown. (d) Ptx does not affect the LPS-induced increment in DC speed (c) but decreases the velocity of LPS-DCs migrating in the presence of CCL21 (d) (positive control for the activity of Ptx, n = 92 and 100 for CCL21 and CCL21 Ptx respectively). 1 representative experiment out of 2 is shown. (e) Mean instantaneous speed of WT and TLR4KO DCs migrating in micro-channels (n = 150, 150, 152 and 122 for iDC, LPS-DC, iTLR4KO and LPS-TLR4KO cells respectively). 1 representative experiment out of 2 is shown. (f) Mean instantaneous speed of control and Myd88KO DCs migrating in micro-channels (n = 81, 74, 67 and 102 for iDC, LPS-DC, iMyd88KO and LPS-Myd88KO respectively). 1 representative experiment out of 3 is shown. The Mann-Whitney test was applied for statistical analyses in c–f. In the box plots of panels c–f the bars include 90% of the points, the center corresponds to the median and the box contains 75% of the data.
Supplementary Figure 2 Quantification of LifeAct-GFP dynamics in migrating DCs.
(a–b) Method used to quantify LifeAct-GFP localization in migrating DCs. Sequential images of LifeAct-GFP DCs were acquired on an epifluorescence microscope every 1 min at 20×. Scale bar: 10 μm. (a) The LifeAct-GFP signal recorded at each time-point was integrated into a single image for all migrating cells. Cell alignment and cell size normalization were applied to generate the LifeAct-GFP density maps. Scale bar: 5 μm. (b) Mean intensities obtained for each cell were averaged into a single LifeAct-GFP density map, assigning equivalent weight to each cell. Scale bar: 2.5 μm. (c,d) Analysis of the LifeAct-GFP front/back ratio in iDCs and LPS-DCs migrating in micro-channels. The front was defined as the first third of the cell. Scale bar: 2.5 μm. Results obtained by analyzing the data showed in Fig. 2a–c (n = 31 and 27 cells for iDC and LPS-DC respectively). 1 representative experiment out of 4 is shown. Graphic shows mean and error bars correspond to S.E.M. The Mann-Whitney test was applied for all statistical analysis.
Supplementary Figure 3 Arp2/3 decreases the migration of iDCs but enhances their antigen uptake capacity.
(a) Immunoblot blot analysis of Arpc4 expression in BMDCs nucleofected with Arpc4-specific siRNA. (b,c) LifeAct-GFP density maps obtained from iDC or LPS-DC silenced (b) or inhibited (c) for Arp2/3. LPS-DCs were treated with the Arp2/3 inhibitor CK666 (25 μM) or silenced for Arpc4. Scale bar: 2.5 μm. 1 representative experiment out of 2 is shown. (d) Analysis by flow cytometry of surface expression of the DC activation marker CD86. iDCs were incubated 30 min with LPS (100 ng/ml), washed and cultured overnight. 1 experiment out of 4 is shown. (e) Mean instantaneous speed of DCs migrating in micro-channels and treated or not with CK666 as in c (n = 244, 192, 231 and 230 for iDC, iDC-CK666, LPS-DC and LPS-DC CK666 respectively). 1 representative experiment out of 3 is shown. (f) Immunoblot analysis of Arpc2 and tubulin expression in tamoxifen-treated DCs. TomatoFP+ (Arpc2WT) and conditional GFP+ (Arpc2KO) iDCs were sorted from 10-days- DC cultures in which tamoxifen was added during the last 8 days. (g) Analysis of the migration of Arpc2WT and KO iDCs under an agarose gel. Cells were imaged during 200 min and their cell speed was quantified after tracking. The analysis was performed on cells that had migrated >100 μm (n = 80 and 163 for Arpc2 WT and Arpc2 KO respectively). 1 representative experiment out of 2 is shown. (h) Analysis by flow cytometry of surface expression of the DC activation marker CD86 performed as in d. (i) Quantification of the size (left) and number (right) of vesicles present at the front of Arpc2WT and KO DCs migrating in micro-channels filled with fluorescent ovalbumin (n = 36 and 37 for Arpc2WT and Arpc2 KO respectively). Graphics show mean and error bars correspond to S.E.M. (j) Macropinocytic capacity of migrating DCs treated with the Arp2/3 inhibitor CK666 (n = 62 and 41 for iDC and iDC CK666 respectively). 1 representative experiment out of 3 is shown. (k) Macropinocytic capacity of iDCs and LPS-DCs (n = 39 and 33 for iDC and LPS-DC respectively). 1 representative experiment out of 2 is shown. (l) Density maps obtained from LifeAct-GFP DCs fixed while migrating in micro-channels and stained for Arp2 and GFP. Scale bar: 2.5 μm. In the box plots of panels e,g,j and k the bars include 90% of the points, the center corresponds to the median and the box contains 75% of the data. The Mann-Whitney test was applied for all statistical analyses.
Supplementary Figure 4 Actin distribution at the back of DCs relies on the Formin mDia1 and is required for fast and persistent migration.
(a) LifeAct-GFP density maps of iDCs migrating in the presence or absence of the formin inhibitor Smifh2 (25 μM). 1 representative experiment out of 3 is shown. Scale bar: 2.5 μm. (b) Immunofluorescence analysis of mDia1 expression in LPS-DCs migrating in micro-channels analyzed by spinning disk microscope (100×). Scale bar: 5 μm. (c) Density maps obtained from LifeAct-GFP DC fixed while migrating in micro-channels and stained for mDia1 and GFP (2 independent experiments). Scale bar: 2.5 μm. (d) Quantitative PCR analysis of mDia1 expression in DCs nucleofected with mDia1-specific siRNA. Graphic shows mean and error bars correspond to S.E.M. (e) Immunoblot analysis of mDia1 and tubulin expression in immature mDia1WT and KO DCs. 1 representative experiment out of 3 is shown. (f) Mean instantaneous speed of LPS-DCs migrating in micro-channels and treated with different doses of Smifh2 (n = 125, 154, 134 and 141 for iDC, LPS-DC, LPS-DC 12 μM and LPS-DC 25 μM respectively). 1 representative experiment out of 3 is shown. (g) 10% of the highest instantaneous speed values of DCs migrating in micro-channels (n = 210, 238, 233 and 179 for iWT, LPS-WT, imDia1KO and LPS-mDia1KO respectively). 1 representative experiment out of 5 is shown. (h) Analysis by flow cytometry of surface expression of the DC activation marker CD86. iDCs were incubated 30 min with LPS (100 ng/ml), washed and cultured overnight. 1 experiment out of 4 is shown. (i) Mean instantaneous speed of mDia1WT and KO migrating in micro-channels and treated with CK666 (25 μM) (n = 119, 107, 52 and 111 for iWT, iWT CK, imDia1KO and imDia1KO CK respectively). 1 representative experiment out of 2 is shown. In the box plots of panels f, g and i the bars include 90% of the points, the center corresponds to the median and the box contains 75% of the data. The Mann-Whitney test was applied for all statistical analyses.
Supplementary Figure 5 Regulation of DC Arp2/3- and mDia1-dependent actin nucleation by Cdc42 and RhoA.
(a, b) iDCs were treated with LPS for 30 min, washed and further cultured at 37 °C during 16 h. a. Immunoblot analysis was performed as described in the supplementary experimental procedures. (b) Normalized intensity obtained from data showed in panel a. Graphics show mean individual values. (c) Mean instantaneous speed of iDCs migrating in micro-channels and treated with different doses of ML141 (n = 164, 163, 142, 143 and 39 for iDC, iDC 12 μM, iDC 25 μM, iDC 50 μM and iDC 100 μM respectively). 1 experiment out of 3 is shown. In the box plot the bars include 90% of the points, the center corresponds to the median and the box contains 75% of the data. The Mann-Whitney test was applied for statistical analysis. (d) Analysis by flow cytometry of surface expression of the DC activation marker CD86. iDCs were incubated 30 min with LPS (100 ng/ml), washed and cultured overnight.
Supplementary Figure 6 Analysis of chemokine gradient sensing by iDCs and LPS-DCs.
(a) Collagen gels were bathed with a solution containing fluorescent ovalbumin (OVA), to evaluate protein penetration in the gel. Gradient steepness was evaluated by measuring the changes in fluorescence intensity according to the protein source using an epifluorescence microscope. Areas closer to the protein source (red) reached a plateau faster than distal areas (blue). After 200 min the gradient was considered as stable at any distance from the source. All the images showed in the paper correspond to analysis between 200 min and 600 min after addition of the chemokine. (b) CCR7 and Clec-2 surface staining using flow cytometry was performed in DCs treated with LPS (100 ng/ml) for 30 min and further cultured overnight. Geometric mean of fluorescence in the CCR7 positive population is depicted. (c) Trajectories of iDCs (upper panel) and LPS-DCs (lower panel) along the CCL21 gradient formed in collagen gels. Directionality of segments of cell trajectories is shown in different colors: from red (toward the source) to blue (against it).
Supplementary Figure 7 Analysis of chemokine gradient sensing by mDia1KO LPS-DCs.
(a) Trajectories of WT (upper panel) and mDia1KO LPS-DCs (lower panel) along the CCL21 gradient formed in collagen gels. Speed of segments of cell trajectories is shown in different colors: from red (fast) to blue (slow). (b) Frequency of movement in the direction of the gradient in the 500 μm closer to the CCL21 source compared to random migration in absence of chemokines. Values are compared to equivalent position in the absence of chemokines. Analysis performed in 500 random tracks due to oversampling. (c) Mean speed of DCs in the 500 μm closer to the CCL21 source. Analysis performed in 500 random tracks due to oversampling. Error bars correspond to S.D. Student’s t-Test was applied for statistical analysis in b. Chi-squared Test was applied for statistical analysis in c. (d) Ear explant stained with the anti-Lyve-1 marker to highlight LVs. The area enclosed in the yellow square corresponds to the zone analyzed when monitoring DC migration to LVs. Scale bar 250 μm. (e) Scheme of different types of trajectories representing the behavior of mDia1WT and KO LPS-DCs in the ear explants. (f) mDia1WT and KO LPS-DCs were stained with CMTRM or CFSE and co-injected in the footpad of WT recipient mice. Their presence in the popliteal lymph node 16 h later was analyzed by flow cytometry. Example representative of 3 experiments is shown. (g) pMLC levels on DCs activated 30 min with LPS and cultured during the indicated time. The pMLC signal was normalized to tubulin levels.
Supplementary Figure 8 Unprocessed full scans of key blots.
(a) Full scans of immunoblots showed in Supplementary Fig. 5a. The whole membrane was divided in two based on the 100 kDa molecular weight marker (left panel). Each blot was sequentially incubated with the indicated primary antibody and the corresponding HRP-conjugated secondary antibody (central panel). For loading controls, the membrane containing the 50 kDa band was incubated sequentially with an anti-tubulin antibody and revealed with the corresponding HRP-conjugated secondary antibody (right panel). (b) Full scans of immunoblots showed in Supplementary Fig. 7g. Blots were obtained as described in a, except that the gel was cut based on the 37 kDa molecular weight marker, allowing detection of phospho-MLC and tubulin in the same run.
Supplementary information
Supplementary Information
Supplementary Information (PDF 3794 kb)
Distinct LifeAct-GFP dynamics in iDCs and LPS-DCs.
LifeAct-GFP-expressing DCs migrating in 8 μm × 5 μm fibronectin-coated micro-channels and time-lapsed on a spinning disk microscope (20×) at a frequency of 1 image/2 min. Scale bar 5 μm. (AVI 83 kb)
F-actin is enriched in ruffles at the front of iDCs.
LifeAct-GFP-expressing iDCs migrating in 8 μm × 5 μm fibronectin-coated micro-channels and time-lapsed on a spinning disk microscope (100×) at a frequency of 1 image/400 ms. The cortex of a representative cell is shown. Scale bar 5 μm. (AVI 1988 kb)
F-actin is enriched at the rear of LPS-DCs.
LifeAct-GFP-expressing LPS-DCs migrating in 8 μm × 5 μm fibronectin-coated micro-channels and time-lapsed on a spinning disk microscope (100×) at a frequency of 1 image/400 ms. The cortex of a representative cell is shown. Scale bar 5 μm. (AVI 948 kb)
LPS-DCs but not iDCs respond to CCL21 gradients.
iDCs and LPS-DC migrating along a CCL21 gradient in a collagen gel. The frequency of acquisition was 1 image/2 min (10×) on a video-microscope. The source of CCL21 is at the right of the movie. Scale bar 50 μm. (AVI 5470 kb)
Chemotaxis of wild-type and mDia1KO LPS-DCs in vitro.
mDia1 wild-type and KO LPS-DCs migrating along a CCL21 gradient in a collagen gel. 1 image/2 min (10×) was acquired on a video-microscope. The source of CCL21 is at the right of the movie. Scale bar 50 μm. (AVI 7071 kb)
Chemotaxis of wild-type and mDia1KO LPS-DCs in vivo.
mDia1 wild-type (red) and KO (green) LPS-DCs migrating in an ear explant. 1 image/2 min (20×) was acquired on a on a video-microscope. Scale bar 50 μm. (AVI 3012 kb)
Rights and permissions
About this article
Cite this article
Vargas, P., Maiuri, P., Bretou, M. et al. Innate control of actin nucleation determines two distinct migration behaviours in dendritic cells. Nat Cell Biol 18, 43–53 (2016). https://doi.org/10.1038/ncb3284
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb3284
This article is cited by
-
Increased cross-presentation by dendritic cells and enhanced anti-tumour therapy using the Arp2/3 inhibitor CK666
British Journal of Cancer (2023)
-
Dendritic cell migration in inflammation and immunity
Cellular & Molecular Immunology (2021)
-
The principles of directed cell migration
Nature Reviews Molecular Cell Biology (2021)
-
Sliding walls: a new paradigm for fluidic actuation and protocol implementation in microfluidics
Microsystems & Nanoengineering (2020)
-
Molecular mechanisms of dendritic cell migration in immunity and cancer
Medical Microbiology and Immunology (2020)