Abstract
Cell contacts provide spatial cues that polarize early embryos and epithelial cells. The homophilic adhesion protein E-cadherin is required for contact-induced polarity in many cells. However, it is debated whether E-cadherin functions instructively as a spatial cue, or permissively by ensuring adequate adhesion so that cells can sense other contact signals. In Caenorhabditis elegans, contacts polarize early embryonic cells by recruiting the RhoGAP PAC-1 to the adjacent cortex, inducing PAR protein asymmetry. Here we show that the E-cadherin HMR-1, which is dispensable for adhesion, functions together with the α-catenin HMP-1, the p120 catenin JAC-1, and the previously uncharacterized linker PICC-1 (human CCDC85A-C) to bind PAC-1 and recruit it to contacts. Mislocalizing the HMR-1 intracellular domain to contact-free surfaces draws PAC-1 to these sites and depolarizes cells, demonstrating an instructive role for HMR-1 in polarization. Our findings identify an E-cadherin-mediated pathway that translates cell contacts into cortical polarity by directly recruiting a symmetry-breaking factor to the adjacent cortex.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Roignot, J., Peng, X. & Mostov, K. Polarity in mammalian epithelial morphogenesis. Cold Spring Harb. Perspect. Biol. 5, a013789 (2013).
St Johnston, D. & Ahringer, J. Cell polarity in eggs and epithelia: parallels and diversity. Cell 141, 757–774 (2010).
Johnson, M. H. From mouse egg to mouse embryo: polarities, axes, and tissues. Annu. Rev. Cell Dev. Biol. 25, 483–512 (2009).
Nelson, W. J., Dickinson, D. J. & Weis, W. I. Roles of cadherins and catenins in cell–cell adhesion and epithelial cell polarity. Prog. Mol. Biol. Transl. Sci. 116, 3–23 (2013).
Chen, J. & Zhang, M. The Par3/Par6/aPKC complex and epithelial cell polarity. Exp. Cell Res. 319, 1357–1364 (2013).
Nance, J. & Zallen, J. A. Elaborating polarity: PAR proteins and the cytoskeleton. Development 138, 799–809 (2011).
Adams, C. L., Nelson, W. J. & Smith, S. J. Quantitative analysis of cadherin-catenin-actin reorganization during development of cell–cell adhesion. J. Cell Biol. 135, 1899–1911 (1996).
Adams, C. L., Chen, Y. T., Smith, S. J. & Nelson, W. J. Mechanisms of epithelial cell–cell adhesion and cell compaction revealed by high-resolution tracking of E-cadherin-green fluorescent protein. J. Cell Biol. 142, 1105–1119 (1998).
Vasioukhin, V., Bauer, C., Yin, M. & Fuchs, E. Directed actin polymerization is the driving force for epithelial cell–cell adhesion. Cell 100, 209–219 (2000).
Nejsum, L. N. & Nelson, W. J. A molecular mechanism directly linking E-cadherin adhesion to initiation of epithelial cell surface polarity. J. Cell Biol. 178, 323–335 (2007).
Johnson, M. H., Maro, B. & Takeichi, M. The role of cell adhesion in the synchronization and orientation of polarization in 8-cell mouse blastomeres. J. Embryol. Exp. Morphol. 93, 239–255 (1986).
Gumbiner, B., Stevenson, B. & Grimaldi, A. The role of the cell adhesion molecule uvomorulin in the formation and maintenance of the epithelial junctional complex. J. Cell Biol. 107, 1575–1587 (1988).
McNeill, H., Ozawa, M., Kemler, R. & Nelson, W. J. Novel function of the cell adhesion molecule uvomorulin as an inducer of cell surface polarity. Cell 62, 309–316 (1990).
Watabe, M., Nagafuchi, A., Tsukita, S. & Takeichi, M. Induction of polarized cell–cell association and retardation of growth by activation of the E-cadherin-catenin adhesion system in a dispersed carcinoma line. J. Cell Biol. 127, 247–256 (1994).
Capaldo, C. T. & Macara, I. G. Depletion of E-cadherin disrupts establishment but not maintenance of cell junctions in Madin-Darby canine kidney epithelial cells. Mol. Biol. Cell 18, 189–200 (2007).
Stephenson, R. O., Yamanaka, Y. & Rossant, J. Disorganized epithelial polarity and excess trophectoderm cell fate in preimplantation embryos lacking E-cadherin. Development 137, 3383–3391 (2010).
Nejsum, L. N. & Nelson, W. J. Epithelial cell surface polarity: the early steps. Front. Biosci. 14, 1088–1098 (2009).
Yeaman, C., Grindstaff, K. K. & Nelson, W. J. Mechanism of recruiting Sec6/8 (exocyst) complex to the apical junctional complex during polarization of epithelial cells. J. Cell Sci. 117, 559–570 (2004).
Harris, T. J. & Peifer, M. Adherens junction-dependent and -independent steps in the establishment of epithelial cell polarity in Drosophila. J. Cell Biol. 167, 135–147 (2004).
Le Bivic, A. E-cadherin-mediated adhesion is not the founding event of epithelial cell polarity in Drosophila. Trends Cell Biol. 15, 237–240 (2005).
Costa, M. et al. A putative catenin-cadherin system mediates morphogenesis of the Caenorhabditis elegans embryo. J. Cell Biol. 141, 297–308 (1998).
Tanentzapf, G., Smith, C., McGlade, J. & Tepass, U. Apical, lateral, and basal polarization cues contribute to the development of the follicular epithelium during Drosophila oogenesis. J. Cell Biol. 151, 891–904 (2000).
Theard, D., Steiner, M., Kalicharan, D., Hoekstra, D. & van Ijzendoorn, S. C. Cell polarity development and protein trafficking in hepatocytes lacking E-cadherin/β-catenin-based adherens junctions. Mol. Biol. Cell 18, 2313–2321 (2007).
Nance, J. & Priess, J. R. Cell polarity and gastrulation in C. elegans. Development 129, 387–397 (2002).
Nance, J., Munro, E. M. & Priess, J. R. C. elegans PAR-3 and PAR-6 are required for apicobasal asymmetries associated with cell adhesion and gastrulation. Development 130, 5339–5350 (2003).
Vinot, S. et al. Asymmetric distribution of PAR proteins in the mouse embryo begins at the 8-cell stage during compaction. Dev. Biol. 282, 307–319 (2005).
Thomas, F. C. et al. Contribution of JAM-1 to epithelial differentiation and tight-junction biogenesis in the mouse preimplantation embryo. J. Cell Sci. 117, 5599–5608 (2004).
Plusa, B. et al. Downregulation of Par3 and aPKC function directs cells towards the ICM in the preimplantation mouse embryo. J. Cell Sci. 118, 505–515 (2005).
Anderson, D. C., Gill, J. S., Cinalli, R. M. & Nance, J. Polarization of the C. elegans embryo by RhoGAP-mediated exclusion of PAR-6 from cell contacts. Science 320, 1771–1774 (2008).
Grana, T. M., Cox, E. A., Lynch, A. M. & Hardin, J. SAX-7/L1CAM and HMR-1/cadherin function redundantly in blastomere compaction and non-muscle myosin accumulation during Caenorhabditis elegans gastrulation. Dev. Biol. 344, 731–744 (2010).
Hirano, S., Nose, A., Hatta, K., Kawakami, A. & Takeichi, M. Calcium-dependent cell–cell adhesion molecules (cadherins): subclass specificities and possible involvement of actin bundles. J. Cell Biol. 105, 2501–2510 (1987).
Chihara, D. & Nance, J. An E-cadherin-mediated hitchhiking mechanism for C. elegans germ cell internalization during gastrulation. Development 139, 2547–2556 (2012).
Korswagen, H. C., Herman, M. A. & Clevers, H. C. Distinct β-catenins mediate adhesion and signalling functions in C. elegans. Nature 406, 527–532 (2000).
Pettitt, J., Cox, E. A., Broadbent, I. D., Flett, A. & Hardin, J. The Caenorhabditis elegans p120 catenin homologue, JAC-1, modulates cadherin-catenin function during epidermal morphogenesis. J. Cell Biol. 162, 15–22 (2003).
Daniel, J. M. & Reynolds, A. B. The tyrosine kinase substrate p120cas binds directly to E-cadherin but not to the adenomatous polyposis coli protein or α-catenin. Mol. Cell. Biol. 15, 4819–4824 (1995).
Markham, N. O. et al. Monoclonal antibodies to DIPA: a novel binding partner of p120-catenin isoform 1. Hybridoma 31, 246–254 (2012).
Markham, N. O. et al. DIPA-family coiled-coils bind conserved isoform-specific head domain of p120-catenin family: potential roles in hydrocephalus and heterotopia. Mol. Biol. Cell 25, 2592–2603 (2014).
Audhya, A. et al. A complex containing the Sm protein CAR-1 and the RNA helicase CGH-1 is required for embryonic cytokinesis in Caenorhabditis elegans. J. Cell Biol. 171, 267–279 (2005).
Chan, E. & Nance, J. Mechanisms of CDC-42 activation during contact-induced cell polarization. J. Cell Sci. 126, 1692–1702 (2013).
Stepniak, E., Radice, G. L. & Vasioukhin, V. Adhesive and signaling functions of cadherins and catenins in vertebrate development. Cold Spring Harb. Perspect. Biol. 1, a002949 (2009).
Cardellini, P., Davanzo, G. & Citi, S. Tight junctions in early amphibian development: detection of junctional cingulin from the 2-cell stage and its localization at the boundary of distinct membrane domains in dividing blastomeres in low calcium. Dev. Dyn. 207, 104–113 (1996).
Muller, H. A. & Hausen, P. Epithelial cell polarity in early Xenopus development. Dev. Dyn. 202, 405–420 (1995).
Fesenko, I. et al. Tight junction biogenesis in the early Xenopus embryo. Mech. Dev. 96, 51–65 (2000).
Baas, A. F. et al. Complete polarization of single intestinal epithelial cells upon activation of LKB1 by STRAD. Cell 116, 457–466 (2004).
Sousa, S. et al. ARHGAP10 is necessary for α-catenin recruitment at adherens junctions and for Listeria invasion. Nat. Cell Biol. 7, 954–960 (2005).
Van Itallie, C. M. et al. Biotin ligase tagging identifies proteins proximal to E-cadherin, including lipoma preferred partner, a regulator of epithelial cell–cell and cell-substrate adhesion. J. Cell Sci. 127, 885–895 (2014).
Mori, N. et al. Ccdc85c encoding a protein at apical junctions of radial glia is disrupted in hemorrhagic hydrocephalus (hhy) mice. Am. J. Pathol. 180, 314–327 (2012).
Frokjaer-Jensen, C. et al. Targeted gene deletions in C. elegans using transposon excision. Nat. Methods 7, 451–453 (2010).
Warming, S., Costantino, N., Court, D. L., Jenkins, N. A. & Copeland, N. G. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 33, e36 (2005).
Sarov, M. et al. A recombineering pipeline for functional genomics applied to Caenorhabditis elegans. Nat. Methods 3, 839–844 (2006).
Zhang, Y., Nash, L. & Fisher, A. L. A simplified, robust, and streamlined procedure for the production of C. elegans transgenes via recombineering. BMC Dev. Biol. 8, 119 (2008).
D’Agostino, I., Merritt, C., Chen, P. L., Seydoux, G. & Subramaniam, K. Translational repression restricts expression of the C. elegans Nanos homolog NOS-2 to the embryonic germline. Dev. Biol. 292, 244–252 (2006).
Sarov, M. et al. A genome-scale resource for in vivo tag-based protein function exploration in C. elegans. Cell 150, 855–866 (2012).
Praitis, V., Casey, E., Collar, D. & Austin, J. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics 157, 1217–1226 (2001).
Mello, C. C., Kramer, J. M., Stinchcomb, D. & Ambros, V. Efficient gene transfer in C. elegans extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10, 3959–3970 (1991).
Kamath, R. S., Martinez-Campos, M., Zipperlen, P., Fraser, A. G. & Ahringer, J. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2, RESEARCH0002 (2001).
Totong, R., Achilleos, A. & Nance, J. PAR-6 is required for junction formation but not apicobasal polarization in C. elegans embryonic epithelial cells. Development 134, 1259–1268 (2007).
Kamath, R. S. & Ahringer, J. Genome-wide RNAi screening in Caenorhabditis elegans. Methods 30, 313–321 (2003).
Timmons, L. & Fire, A. Specific interference by ingested dsRNA. Nature 395, 854 (1998).
Schonegg, S. & Hyman, A. A. CDC-42 and RHO-1 coordinate acto-myosin contractility and PAR protein localization during polarity establishment in C. elegans embryos. Development 133, 3507–3516 (2006).
Hadwiger, G., Dour, S., Arur, S., Fox, P. & Nonet, M. L. A monoclonal antibody toolkit for C. elegans. PLoS ONE 5, e10161 (2010).
Shelton, C. A. & Bowerman, B. Time-dependent responses to glp-1-mediated inductions in early C. elegans embryos. Development 122, 2043–2050 (1996).
Acknowledgements
We thank A. Fisher (University of Pittsburgh, USA), K. Gieseler (Université Claude Bernard Lyon 1, France), G. Hermann (Lewis and Clark College, USA), O. Hobert (Columbia University Medical Center, USA), J. Hubbard (New York University School of Medicine, USA), T. Hyman (Max Planck Institute for Molecular Cell Biology and Genetics, Germany) and E. Jorgensen (University of Utah, USA) for their generous gifts of strains, plasmids or antibodies. Special thanks to M. Burel for help in examining GFP–PAC-1ΔPH localization. Thanks to L. Christiaen, T. Hurd, and members of the Nance laboratory for comments on the manuscript. This study was financially supported by a National Science Foundation Graduate Research Fellowship under grant No. 12-A0-00-000165-01 (D.K.), an American Heart Association postdoctoral fellowship (Y.Z.), and NIH grants R01GM098492 (J.N.), R01GM078341 (J.N.) and T32HD007520 (D.C.A.).
Author information
Authors and Affiliations
Contributions
D.K., D.C.A. and J.N. designed, executed and analysed the experiments. J.Y.Y., Y.Z. and J.N. designed, executed and analysed the co-immunoprecipitation experiments. D.K. and J.N. wrote the manuscript with input from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 3 GFP-PAC-1 transgene expression levels and controls for GFP-PAC-1N localization in the absence of endogenous PAC-1.
(a) Expression levels of GFP-PAC-1 transgenes and derivatives, measured by fluorescence intensity (after background subtraction) at the four-cell stage (see Methods), expressed in arbitrary fluorescence units. PAC-1 amino acids present in each GFP fusion protein are indicated (ΔPH lacks the PH domain only). The box represents first and third quartiles, bars represent maximum and minimum values, and the line within the box is the mean fluorescence intensity (full length n = 11 embryos, ΔPH n = 13, 392-838 n = 11, 575-1604 n = 13, 2-610 n = 12, 1-574 n = 14). While expression levels vary between transgenes, contact localization is evident even from those with the lowest expression levels (ex. full-length GFP-PAC-1 and GFP-PAC-12–610). Therefore, lack of apparent contact localization for GFP-PAC-1392–838 and GFP-PAC-1575–1604 is not due to low expression level. (b) Full-length GFP-PAC-1 in control (empty vector) RNAi embryos localizes to cell contacts (arrow, 20/20 embryos). (c) In pac-1(3′ RNAi) embryos GFP-PAC-1 is not detected (19/19 embryos), indicating that RNAi effectively depletes PAC-1. Scale bars, 10 μm.
Supplementary Figure 4 Rescue of pac-1 polarity defects by GFP-PAC-1ΔPH.
(a) Wild-type, (b) pac-1(xn6), and (c) pac-1(xn6); GFP-PAC-1ΔPH 6–8 cell embryos stained for PAR-6. (d) PAR-6 asymmetry is lost in pac-1 mutants but is rescued by expression of GFP-PAC-1ΔPH. PAR-6 asymmetry was quantified by determining the polarity index, defined as the ratio of PAR-6 levels at contact-free surfaces versus half of the PAR-6 level at cell contacts (see Methods). Circles represent individual data points and the gray line indicates the average (wild type n = 8 embryos, pac-1 n = 10, pac-1; GFP-PAC-1ΔPHn = 8). Measured by Mann–Whitney U-test, pac-1(xn6) embryos have a polarity index significantly lower than wild type (∗∗∗p < 0.001) and pac-1(xn6); GFP-PAC-1ΔPH (∗∗∗p < 0.001). The polarity index of pac-1(xn6); GFP-PAC-1ΔPH is marginally significantly different from wild type (∗p < 0.05). Samples were pooled from two independent experiments. Scale bars, 10 μm.
Supplementary Figure 5 Comparison of GFP-PAC-1N in hmp-2(RNAi) and hmp-1(RNAi) embryos.
(a–c) GFP-PAC-1N in live embryos of the indicated genotype; images were captured at the same exposure. (d) Quantification of GFP-PAC-1N contact enrichment. Circles represent data points from individual embryos, and the average is a gray line. ∗∗p < 0.01; n.s. = not significantly different, Mann–Whitney U test. (Control n = 13 embryos, hmp-2(RNAi) n = 9, hmp-1(RNAi) n = 9). Scale bars, 10 μm.
Supplementary Figure 6 Genetic requirements for catenin localization.
Control embryos are wild-type embryos fed on bacteria containing empty RNAi vector. (a,b) HMP-1 immunostaining at cell contacts in control embryos (40/40 embryos) or in the cytoplasm of hmp-2(RNAi) embryos (35/35 embryos). (c,d) α-JAC-1 antiserum immunostains cell contacts in wild-type embryos (30/30 embryos) but not in jac-1(xn15) embryos (42/42 embryos). α-JAC-1 antiserum also stains nuclei non-specifically, as evidenced by the loss of contact staining, but not nuclear staining, in jac-1 mutant embryos (d). (e,f) GFP-JAC-1 at cell contacts in a live control embryo (41/41 embryos) and in the cytoplasm in a live hmr-1 embryo (25/25 embryos). (g) HMP-1 immunostaining in a hmp-1(RNAi) embryo. (h) HMP-1 immunostaining in a hmr-1 embryo, where HMP-1 localizes to the cytoplasm (45/45 embryos). (i) Schematic of the jac-1 gene, with exons shown as rectangles and introns as chevrons. Region encoding the Armadillo (Arm) repeats, which contains the peptide used to generate α-JAC-1 antiserum, is indicated. The dashed region indicates the extent of jac-1 deleted in the xn15 allele. Scale bars, 10 μm.
Supplementary Figure 7 Catenin and HMR-1 localization following catenin depletion.
Control embryos are wild-type embryos fed on bacteria containing RNAi empty vector. (a,b) HMP-1-GFP at cell contacts in wild-type and jac-1 embryos. (c) Quantification of HMP-1-GFP contact enrichment in wild-type and jac-1 embryos (wild type n = 12 embryos, jac-1 n = 12). Circles represent individual values and gray lines the mean. HMP-1-GFP levels at contacts in the two genotypes are not significantly different (p = 0.06, Mann–Whitney U test). (d,e) GFP-JAC-1 at cell contacts in a control and hmp-1(RNAi) embryo. (f) Quantification of GFP-JAC-1 contact enrichment in control and hmp-1(RNAi) embryos. (control n = 13 embryos, hmp-1(RNAi) n = 12). GFP-JAC-1 levels at contacts in the two genotypes are not significantly different (p = 0.3, Mann–Whitney U test). (g–j) HMR-1-GFP expression in live four-cell embryos of the indicated genotype. (k) Quantification of HMR-1-GFP levels at cell contacts in embryos of the indicated genotype (control n = 18 embryos, hmp-1(RNAi) n = 33, jac-1 n = 17, jac-1 + hmp-1(RNAi)n = 17). None of the mean values are significantly different from control (p > 0.05, Mann–Whitney U test). Control samples and hmp-1(RNAi) embryos were pooled from two independent experiments. (l,m) Endogenous HMR-1 immunostaining at cell contacts in control (91/91 embryos) and jac-1(xn15) + hmp-1(RNAi) (133/133 embryos). Scale bars, 10 μm.
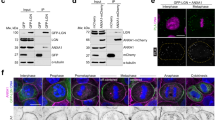
Supplementary Figure 8 picc-1 gene and regulation of PICC-1 localization.
(a) Schematic of the picc-1 gene, with exons shown as rectangles and introns as chevrons. The dashed region indicates portion of the picc-1 gene deleted in the xn14 allele. (b) Region of conservation between PICC-1 and homologues in human (CCDC85A-C; CCDC85B is also known as DIPA) and Drosophila (CG17265). Black shading indicates three or more identical residues; gray shading indicates similar residues. (c,d) GFP-PICC-1 localization in a hmr-1 mutant embryo (n = 50 embryos) and a hmp-1(RNAi) embryo (n = 31 embryos). Compare to wild-type localization in Figure 4a. Scale bars, 10 μm.
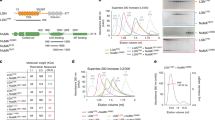
Supplementary Figure 9 Two-hybrid controls and reciprocal immunoprecipitations.
(a) Control two-hybrid experiments, performed as described in Figure 5 legend. (b) Immunoprecipitation of PICC-1-GFP from embryonic lysates showing co-immunoprecipitation of JAC-1 species. The experiment was performed seven times, and a representative example is shown. (c) Immunoprecipitation of mCherry-HA-PAC-1 from embryonic lysates showing co-immunoprecipitation of PICC-1-GFP. The experiment was performed four times, and a representative example is shown. Tubulin levels in the input (total embryonic lysate) are shown as a loading control. Uncropped blots are shown in Supplementary Figure 8.
Supplementary information
Supplementary Information
Supplementary Information (PDF 690 kb)
Rights and permissions
About this article
Cite this article
Klompstra, D., Anderson, D., Yeh, J. et al. An instructive role for C. elegans E-cadherin in translating cell contact cues into cortical polarity. Nat Cell Biol 17, 726–735 (2015). https://doi.org/10.1038/ncb3168
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb3168
This article is cited by
-
Computational study of biomechanical drivers of renal cystogenesis
Biomechanics and Modeling in Mechanobiology (2023)
-
Apical–basal polarity and the control of epithelial form and function
Nature Reviews Molecular Cell Biology (2022)
-
Beyond β-catenin: prospects for a larger catenin network in the nucleus
Nature Reviews Molecular Cell Biology (2016)