Abstract
Although recent studies have shown that adenosine-to-inosine (A-to-I) RNA editing occurs in microRNAs (miRNAs), its effects on tumour growth and metastasis are not well understood. We present evidence of CREB-mediated low expression of ADAR1 in metastatic melanoma cell lines and tumour specimens. Re-expression of ADAR1 resulted in the suppression of melanoma growth and metastasis in vivo. Consequently, we identified three miRNAs undergoing A-to-I editing in the weakly metastatic melanoma but not in strongly metastatic cell lines. One of these miRNAs, miR-455-5p, has two A-to-I RNA-editing sites. The biological function of edited miR-455-5p is different from that of the unedited form, as it recognizes a different set of genes. Indeed, wild-type miR-455-5p promotes melanoma metastasis through inhibition of the tumour suppressor gene CPEB1. Moreover, wild-type miR-455 enhances melanoma growth and metastasis in vivo, whereas the edited form inhibits these features. These results demonstrate a previously unrecognized role for RNA editing in melanoma progression.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Siegel, R., Ma, J., Zou, Z. & Jemal, A. Cancer statistics, 2014. CA Cancer J. Clin. 64, 9–29 (2014).
Miller, A. J. & Mihm, M. C. Jr Melanoma. N. Engl. J. Med. 355, 51–65 (2006).
Xie, S. et al. Dominant-negative CREB inhibits tumor growth and metastasis of human melanoma cells. Oncogene 15, 2069–2075 (1997).
Mobley, A. K., Braeuer, R. R., Kamiya, T., Shoshan, E. & Bar-Eli, M. Driving transcriptional regulators in melanoma metastasis. Cancer Metastasis Rev. 31, 621–632 (2012).
Jean, D., Harbison, M., McConkey, D. J., Ronai, Z. & Bar-Eli, M. CREB and its associated proteins act as survival factors for human melanoma cells. J. Biol. Chem. 273, 24884–24890 (1998).
Melnikova, V. O. & Bar-Eli, M. Transcriptional control of the melanoma malignant phenotype. Cancer Biol. Ther. 7, 997–1003 (2008).
Melnikova, V. O., Mourad-Zeidan, A. A., Lev, D. C. & Bar-Eli, M. Platelet-activating factor mediates MMP-2 expression and activation via phosphorylation of cAMP-response element-binding protein and contributes to melanoma metastasis. J. Biol. Chem. 281, 2911–2922 (2006).
Dobroff, A. S. et al. Silencing cAMP-response element-binding protein (CREB) identifies CYR61 as a tumor suppressor gene in melanoma. J. Biol. Chem. 284, 26194–26206 (2009).
Bertolotto, C. et al. Microphthalmia gene product as a signal transducer in cAMP-induced differentiation of melanocytes. J. Cell Biol. 142, 827–835 (1998).
Melnikova, V. O. et al. CREB inhibits AP-2α expression to regulate the malignant phenotype of melanoma. PLoS ONE 5, e12452 (2010).
Venables, J. P. Unbalanced alternative splicing and its significance in cancer. Bioessays 28, 378–386 (2006).
David, C. J. & Manley, J. L. Alternative pre-mRNA splicing regulation in cancer: pathways and programs unhinged. Genes Dev. 24, 2343–2364 (2010).
Gallo, A. & Galardi, S. A-to-I RNA editing and cancer: from pathology to basic science. RNA Biol. 5, 135–139 (2008).
Levanon, E. Y. et al. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat. Biotechnol. 22, 1001–1005 (2004).
Laurencikiene, J., Kallman, A. M., Fong, N., Bentley, D. L. & Ohman, M. RNA editing and alternative splicing: the importance of co-transcriptional coordination. EMBO Rep. 7, 303–307 (2006).
Jin, Y., Zhang, W. & Li, Q. Origins and evolution of ADAR-mediated RNA editing. IUBMB Life 61, 572–578 (2009).
Bass, B. L. RNA editing by adenosine deaminases that act on RNA. Annu. Rev. Biochem. 71, 817–846 (2002).
Keegan, L. P., Gallo, A. & O’Connell, M. A. The many roles of an RNA editor. Nat. Rev. Genet. 2, 869–878 (2001).
Maas, S., Rich, A. & Nishikura, K. A-to-I RNA editing: recent news and residual mysteries. J. Biol. Chem. 278, 1391–1394 (2003).
Hood, J. L. & Emeson, R. B. Editing of neurotransmitter receptor and ion channel RNAs in the nervous system. Curr. Top. Microbiol. Immunol. 353, 61–90 (2012).
Wulff, B. E. & Nishikura, K. Modulation of microRNA expression and function by ADARs. Curr. Top. Microbiol. Immunol. 353, 91–109 (2012).
Zinshteyn, B. & Nishikura, K. Adenosine-to-inosine RNA editing. Wiley Interdiscip. Rev. Syst. Biol. Med. 1, 202–209 (2009).
Heale, B. S. et al. Editing independent effects of ADARs on the miRNA/siRNA pathways. EMBO J. 28, 3145–3156 (2009).
Nemlich, Y. et al. MicroRNA-mediated loss of ADAR1 in metastatic melanoma promotes tumor growth. J. Clin. Invest. 123, 2703–2718 (2013).
Alon, S. et al. Systematic identification of edited microRNAs in the human brain. Genome Res. 22, 1533–1540 (2012).
Si, K. et al. A neuronal isoform of CPEB regulates local protein synthesis and stabilizes synapse-specific long-term facilitation in aplysia. Cell 115, 893–904 (2003).
Sasayama, T. et al. Over-expression of Aurora-A targets cytoplasmic polyadenylation element binding protein and promotes mRNA polyadenylation of Cdk1 and cyclin B1. Genes Cells 10, 627–638 (2005).
Kawahara, Y., Zinshteyn, B., Chendrimada, T. P., Shiekhattar, R. & Nishikura, K. RNA editing of the microRNA-151 precursor blocks cleavage by the Dicer-TRBP complex. EMBO Rep. 8, 763–769 (2007).
Ota, H. et al. ADAR1 forms a complex with Dicer to promote MicroRNA processing and RNA-induced gene silencing. Cell 153, 575–589 (2013).
Yang, W. et al. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat. Struct. Mol. Biol. 13, 13–21 (2006).
Caldeira, J. et al. CPEB1, a novel gene silenced in gastric cancer: a Drosophila approach. Gut 61, 1115–1123 (2012).
Zhang, J. H. et al. Cytoplasmic polyadenylation element binding protein is a conserved target of tumor suppressor HRPT2/CDC73. Cell Death Differ. 17, 1551–1565 (2010).
Levanon, E. Y. et al. Evolutionarily conserved human targets of adenosine to inosine RNA editing. Nucleic Acids Res. 33, 1162–1168 (2005).
Galeano, F. et al. Human BLCAP transcript: new editing events in normal and cancerous tissues. Int. J. Cancer 127, 127–137 (2010).
Gommans, W. M., McCane, J., Nacarelli, G. S. & Maas, S. A mammalian reporter system for fast and quantitative detection of intracellular A-to-I RNA editing levels. Anal. Biochem. 399, 230–236 (2010).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Landen, C. N. Jr et al. Therapeutic EphA2 gene targeting using neutral liposomal small interfering RNA delivery. Cancer Res. 65, 6910–6918 (2005).
Villares, G. J. et al. Targeting melanoma growth and metastasis with systemic delivery of liposome-incorporated protease-activated receptor-1 small interfering RNA. Cancer Res. 68, 9078–9086 (2008).
Acknowledgements
We thank S. Maas from NIH for providing the ADAR1 A-to-I RNA-editing hairpin loop luciferase reporter plasmid. We thank W. Choi for her help with the miRNA microarray. We thank R. Rajesha for his technical help and support. These studies are supported by SINF, an MDACC grant and NIH Skin Cancer SPORE p50 CA093459.
Author information
Authors and Affiliations
Contributions
M.B-E. conceived and supervised the project. E.S. and A.K.M. designed and carried out experiments and analysed most of the data. R.R.B., T.K., L.H., M.E.V., G.V-T. and A.S. carried out experiments and analysed data. H.J.L., S.J.K. and I.J.F. helped with the spontaneous animal model. C.I., A.G.R., K.M.N., K.Z., D.B., S.J.M.J., I.B., M.M., Y-y.W., A.K.E., P.H. and J.E.G. carried out sequencing and analysis. A.K.S., G.A.C. and G.M. helped with miRNA analysis.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 ADAR2 is not regulated by CREB.
Western blot analyses demonstrating that overexpression of CREB in SB2 cells and silencing CREB in C8161 cells did not affect the expression of ADAR2 in these cells. Note a doublet bands in ADAR2 expression in SB2 cells which represent two different isoforms (73,76KDa) (data are representative of 3, biologically independent experiments). Uncropped images of the blots are shown in Supplementary Figure 8.
Supplementary Figure 2 Luciferase imaging of lymph node metastasis in nude mice.
Images of mice after the primary tumors were removed show the presence or absence of local lymph node metastases. These mice represent the results described in Figure 4C (n = 8 mice in each group).
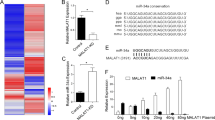
Supplementary Figure 3 Identification of miRs in common between CREB shRNA and ADAR1 shRNA arrays.
(A) Venn diagram showing the breakdown of the differentially regulated miRNAs between the two arrays analyzed. (B) Table of the 12 miRNAs identified to be in common between both the CREB shRNA and the ADAR1 shRNA array.
Supplementary Figure 4 miR-378-3p and miR-324-5p are edited by ADAR1 at one A-to-I editing site.
(A) Mature miR-378 sequence highlighted in green. The arrow indicates the RNA editing site. (B) DNA sequencing data indicates a decrease in A-to-I editing in SB2 cells when ADAR1 is silenced. (C) An increase in RNA editing in C8161 cells is observed when ADAR1 is overexpressed. (D) A-to-I RNA editing is increased on CREB silencing and is returned to NT levels on rescue of CREB in C8161 cells. Editing sites are indicated by a red arrow. (E) Mature miR-324 sequence is highlighted in green. The arrow indicates the RNA editing site. (F) DNA sequencing data indicates a decrease in A-to-I editing in SB2 cells when ADAR1 is silenced. (G) An increase in RNA editing is observed in C8161 cells when ADAR1 is overexpressed. (H) A-to-I RNA editing is increased on CREB silencing and is returned to NT levels on rescue of CREB in C8161 cells. Editing sites are indicated by a red arrow.
Supplementary Figure 5 Bound miR-455 to Drosha and Dicer correlates with ADAR1 expression.
(A) Silencing ADAR1 in SB2 melanoma cells results in increased binding of mir-455 to Dicer, (B) Overexpressing ADAR1 in C8161 melanoma cells reduced the binding of mir-455 to Dicer. (C). Overexpression of ADAR1 in C8161 melanoma cells reduced the amount of pri-miR-455 bound to Drosha. (D) Silencing ADAR1 in SB2 melanoma cells resulted with increased interaction between pri-miR-455 and Drosha (n = 3 biologically independent samples; statistical significance by two-tailed Student t-test; error bars represent s.d., p < 0.05).
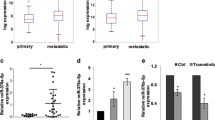
Supplementary Figure 6 Confirmation of manipulation of miR-455-5p levels.
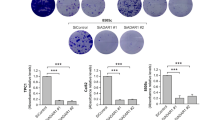
(A) rtPCR analysis of miR-455-5p levels show that wild-type miR-455-5p is overexpressed 2 fold, edited1 miR-455-5p is overexpressed ∼20 fold, edited2 miR-455-5p is overexpressed ∼9 fold and double edited miR-455-5p is overexpressed ∼40 fold, as compared to empty vector control. The antagomir to miR-455-5p shows a downregulation of miR-455-5p by about 90% as compared to the NT control (n = 3 biologically independent samples; statistical significance was determined by two-tailed Student t-test; error bars represent s.d., p < 0.05). (B) Rescue of CPEB1 expression in SB2 ADAR1 KD cells inhibits their invasive potential in matrigel invasion assay, without affecting their proliferation rate (n = 3 biologically independent samples; statistical significance was determined by Tukey’s multiple comparison test; error bars represent s.d., p < 0.05). (C) No change was observed in proliferation rate between SB2 cells transfected with ADAR1-NT, ADAR1-KD, and ADAR1-KD CPEB1-OE (n = 12 biologically independent samples per goup. Statistical significance was determined by one-way ANOVA; error bars represent s.d., p = 0.9610).
Supplementary Figure 7 Potential edited miR-455-5p binding sites on 3'UTR of ITGα2, MDM4 and RhoC.
miR-455-5p seed sequence is depicted in red and the edited sites are shown in green. 6-mer or 7-mer binding sites are highlighted in yellow.
Supplementary Figure 8
Uncropped Western blots and gel images.
Supplementary information
Supplementary Information
Supplementary Information (PDF 1731 kb)
Supplementary Table 1
Supplementary Information (XLSX 19 kb)
Supplementary Table 3
Supplementary Information (XLSX 24 kb)
Rights and permissions
About this article
Cite this article
Shoshan, E., Mobley, A., Braeuer, R. et al. Reduced adenosine-to-inosine miR-455-5p editing promotes melanoma growth and metastasis. Nat Cell Biol 17, 311–321 (2015). https://doi.org/10.1038/ncb3110
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb3110
This article is cited by
-
The role of ADAR1 through and beyond its editing activity in cancer
Cell Communication and Signaling (2024)
-
RNA modification: mechanisms and therapeutic targets
Molecular Biomedicine (2023)
-
Emerging role of the RNA-editing enzyme ADAR1 in stem cell fate and function
Biomarker Research (2023)
-
Biological roles of A-to-I editing: implications in innate immunity, cell death, and cancer immunotherapy
Journal of Experimental & Clinical Cancer Research (2023)
-
RNA modifications in cancer
British Journal of Cancer (2023)