Abstract
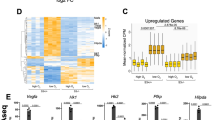
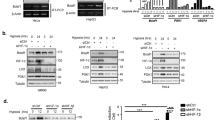
The Hippo signalling pathway plays important roles in animal development, physiology and tumorigenesis1,2,3. Understanding how the activity of this pathway is regulated by the cellular microenvironment remains a major challenge. Here we elucidate a molecular mechanism by which hypoxia deactivates Hippo signalling. We demonstrate that the E3 ubiquitin ligase SIAH2 stimulates YAP by destabilizing LATS2, a critical component of the Hippo pathway, in response to hypoxia. Loss of SIAH2 suppresses tumorigenesis in a LATS2-dependent manner in a xenograft mouse model. We further show that YAP complexes with HIF1α and is essential for HIF1α stability and function in tumours in vivo. LATS2 is downregulated in human breast tumours and negatively correlates with SIAH2 expression levels, indicating that the SIAH2–LATS2 pathway may have a role in human cancer. Our data uncover oxygen availability as a microenvironment signal for the Hippo pathway and have implications for understanding the regulation of Hippo signalling in tumorigenesis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Pan, D. The hippo signaling pathway in development and cancer. Dev. Cell 19, 491–505 (2010).
Harvey, K. F., Zhang, X. & Thomas, D. M. The Hippo pathway and human cancer. Nat. Rev. Cancer 13, 246–257 (2013).
Zhao, B., Tumaneng, K. & Guan, K. L. The Hippo pathway in organ size control, tissue regeneration and stem cell self-renewal. Nat. Cell Biol. 13, 877–883 (2011).
Wu, S., Huang, J., Dong, J. & Pan, D. hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell 114, 445–456 (2003).
Harvey, K. F., Pfleger, C. M. & Hariharan, I. K. The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell 114, 457–467 (2003).
Pantalacci, S., Tapon, N. & Leopold, P. The Salvador partner Hippo promotes apoptosis and cell-cycle exit in Drosophila. Nat. Cell Biol. 5, 921–927 (2003).
Udan, R. S., Kango-Singh, M., Nolo, R., Tao, C. & Halder, G. Hippo promotes proliferation arrest and apoptosis in the Salvador/Warts pathway. Nat. Cell Biol. 5, 914–920 (2003).
Huang, J., Wu, S., Barrera, J., Matthews, K. & Pan, D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila homolog of YAP. Cell 122, 421–434 (2005).
Dong, J. et al. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell 130, 1120–1133 (2007).
Zhao, B. et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 21, 2747–2761 (2007).
Hisaoka, M., Tanaka, A. & Hashimoto, H. Molecular alterations of h-warts/LATS1 tumor suppressor in human soft tissue sarcoma. Lab. Invest. 82, 1427–1435 (2002).
Jimenez-Velasco, A. et al. Downregulation of the large tumor suppressor 2 (LATS2/KPM) gene is associated with poor prognosis in acute lymphoblastic leukemia. Leukemia 19, 2347–2350 (2005).
Ishizaki, K. et al. Frequent polymorphic changes but rare tumor specific mutations of the LATS2 gene on 13q11-12 in esophageal squamous cell carcinoma. Int. J. Oncol. 21, 1053–1057 (2002).
Powzaniuk, M. et al. The LATS2/KPM tumor suppressor is a negative regulator of the androgen receptor. Mol. Endocrinol. 18, 2011–2023 (2004).
Semenza, G. L. Hypoxia-inducible factors in physiology and medicine. Cell 148, 399–408 (2012).
Brown, J. M. & Wilson, W. R. Exploiting tumour hypoxia in cancer treatment. Nat. Rev. Cancer 4, 437–447 (2004).
Nakayama, K. et al. Siah2 regulates stability of prolyl-hydroxylases, controls HIF1alpha abundance, and modulates physiological responses to hypoxia. Cell 117, 941–952 (2004).
Nakayama, K., Qi, J. & Ronai, Z. The ubiquitin ligase Siah2 and the hypoxia response. Mol. Cancer Res. 7, 443–451 (2009).
Qi, J. et al. Siah2-dependent concerted activity of HIF and FoxA2 regulates formation of neuroendocrine phenotype and neuroendocrine prostate tumors. Cancer Cell 18, 23–38 (2010).
Calzado, M. A., de la Vega, L., Moller, A., Bowtell, D. D. & Schmitz, M. L. An inducible autoregulatory loop between HIPK2 and Siah2 at the apex of the hypoxic response. Nat. Cell Biol. 11, 85–91 (2009).
Habelhah, H. et al. Regulation of 2-oxoglutarate (alpha-ketoglutarate) dehydrogenase stability by the RING finger ubiquitin ligase Siah. J. Biol. Chem. 279, 53782–53788 (2004).
Christian, P. A., Fiandalo, M. V. & Schwarze, S. R. Possible role of death receptor-mediated apoptosis by the E3 ubiquitin ligases Siah2 and POSH. Mol. Cancer 10, 57 (2011).
Wong, C. S. & Moller, A. Siah: a promising anticancer target. Cancer Res. 73, 2400–2406 (2013).
Hu, G. et al. Characterization of human homologs of the Drosophila seven in absentia (sina) gene. Genomics 46, 103–111 (1997).
Tao, W. et al. Human homologue of the Drosophila melanogaster lats tumour suppressor modulates CDC2 activity. Nat. Genet. 21, 177–181 (1999).
Yabuta, N. et al. Structure, expression, and chromosome mapping of LATS2, a mammalian homologue of the Drosophila tumor suppressor gene lats/warts. Genomics 63, 263–270 (2000).
Sun, R. C. & Denko, N. C. Hypoxic regulation of glutamine metabolism through HIF1 and SIAH2 supports lipid synthesis that is necessary for tumor growth. Cell Metab. 19, 285–292 (2014).
Shah, M. et al. Inhibition of Siah2 ubiquitin ligase by vitamin K3 (menadione) attenuates hypoxia and MAPK signaling and blocks melanoma tumorigenesis. Pigment Cell Melanoma Res. 22, 799–808 (2009).
Bendinelli, P. et al. Hypoxia inducible factor-1 is activated by transcriptional co-activator with PDZ-binding motif (TAZ) versus WWdomain-containing oxidoreductase (WWOX) in hypoxic microenvironment of bone metastasis from breast cancer. Eur. J. Cancer 49, 2608–2618 (2013).
Matteucci, E. et al. Bone metastatic process of breast cancer involves methylation state affecting E-cadherin expression through TAZ and WWOX nuclear effectors. Eur. J. Cancer 49, 231–244 (2013).
Kaelin, W. G. Jr. & Ratcliffe, P. J. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol. Cell 30, 393–402 (2008).
Epstein, A. C. et al. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell 107, 43–54 (2001).
Saucedo, L. J. & Edgar, B. A. Filling out the Hippo pathway. Nat. Rev. Mol. Cell Biol. 8, 613–621 (2007).
Badouel, C., Garg, A. & McNeill, H. Herding Hippos: regulating growth in flies and man. Curr. Opin. Cell Biol. 21, 837–843 (2009).
Halder, G. & Johnson, R. L. Hippo signaling: growth control and beyond. Development 138, 9–22 (2011).
Irvine, K. D. Integration of intercellular signaling through the Hippo pathway. Semin. Cell Dev. Biol. 23, 812–817 (2012).
Pouyssegur, J., Dayan, F. & Mazure, N. M. Hypoxia signalling in cancer and approaches to enforce tumour regression. Nature 441, 437–443 (2006).
Acknowledgements
We thank H. Deng from Tsinghua University for Mass spectrometry analysis, Y. (YZ) Luo from Tsinghua University for providing the HRE–luciferase reporter plasmid and Y. (YG) Chen from Tsinghua University for providing the HA–VHL plasmid. We are deeply grateful to D. (DJ) Pan from Johns Hopkins University for his suggestions during the manuscript revision. This work was supported by the National Basic Research Program of China (2010CB912204, 2011CB9109903, 2011CB943903 and 2013CB531200) and the National Natural Sciences Foundation of China (31471319, 81130045 and 31171411).
Author information
Authors and Affiliations
Contributions
B.M. conceived and designed the experiments with Y.Z., Q.C. and S.W. B.M. performed most of the experiments and data analysis in the laboratories of Q.C. and Y.Z. B.M., C.M. and R.G. performed xenograft implantation experiments. B.M. and C.M. performed studies on tissue microarrays of human patient samples. L. Chen, H.C. and J. Li contributed to cellular experiments and plasmid construction and protein purification. Y.C., L. Cao, C.Z. and J. Liu provided technical support. B.M. and S.W. wrote the manuscript with the help of all authors. Q.C. initiated, and supervised the project together with Y.Z. and S.W.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 5 Schematic drawing of LATS2 deletion mutants and their responses to interaction with and degradation by SIAH2.
A schematic diagram of LATS2 deletion mutants is shown on the left panel. Blue represents as LATS Conserved Domain 1(LCD1) (aa. 1–160), yellow as LATS Conserved Domain 2(LCD2) (aa. 403–463), red as PAPA repeat (aa. 467–480), green as Protein Binding Domain (PBD) (aa. 618–720), purple as Kinase Domain. LATS2 binding activity to SIAH2 is shown in the middle panel as assayed in Fig. 1d. “+” indicates as binding, “−” as no binding, and “−/+” as less binding. The right panel shows the responses of LATS2 mutants to SIAH2-induced degradation. Cells expressing constant amounts of Myc-LATS2 mutant and increasing amounts of Flag-SIAH2 were subjected to immunoblotting. “+” indicates response to SIAH2-induced degradation and “−” indicates non-response.
Supplementary Figure 6 SIAH2 destabilizes LATS2 through proteasome.
(a,b) LATS2 degradation was dosage-dependent on SIAH2 (a), but not on SIAH2RM, an E3 ligase-activity-dead mutant of SIAH2 (b). (c,d) SIAH2-mediated LATS2 degradation was inhibited by proteasomal inhibitor MG132 (c), but not by lysosomal inhibitor BA-1 (d). (e,f) The half-life of LATS2 was shortened by SIAH2 (e) but not by SIAH2RM (f) in cycloheximide chase assays.
Supplementary Figure 7 LATS1/2 is specifically regulated by SIAH2 but not by SIAH1.
(a) No co-immunoprecipitation of exogenously expressed Myc-LATS2 by Flag-SIAH1RM. (b) LATS2 degradation is specifically promoted by SIAH2 but not by SIAH1. (c) Co-immunoprecipitation of exogenous expressed Flag-SIAH2 by Myc-LATS1 and vice versa. (d) LATS1 stability is regulated by SIAH2 but not by SIAH2RM. (e) No co-immunoprecipitation of exogenously expressed Myc-LATS1 by Flag-SIAH1RM.
Supplementary Figure 8 YAP protects HIF1α from proteasomal degradation under hypoxia.
(a) Scramble and YAP knockdown MDA-MB-231 cells were cultured under hypoxic conditions. Total RNA was extracted and subjected to RT-qPCR analysis for HIF1α mRNA expression. (b) MG132 stabilized HIF1α in YAP knockdown cells under hypoxic conditions. (c) Scramble and YAP knockdown MDA-MB-231 cells were cultured under hypoxic conditions in the presence of MG132. Protein expressions were analysed by immunoblotting with indicated antibodies. Data in a is the mean of n = 3 independent experiments and error bars indicate s.d. Two-tailed, unpaired Student’s t-test.
Supplementary information
Supplementary Information
Supplementary Information (PDF 1485 kb)
Rights and permissions
About this article
Cite this article
Ma, B., Chen, Y., Chen, L. et al. Hypoxia regulates Hippo signalling through the SIAH2 ubiquitin E3 ligase. Nat Cell Biol 17, 95–103 (2015). https://doi.org/10.1038/ncb3073
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb3073
This article is cited by
-
LATS2 degradation promoted fibrosis damage and rescued by vitamin K3 in lupus nephritis
Arthritis Research & Therapy (2024)
-
New insights into the ambivalent role of YAP/TAZ in human cancers
Journal of Experimental & Clinical Cancer Research (2023)
-
Targeting tumor-stroma communication by blocking endothelin-1 receptors sensitizes high-grade serous ovarian cancer to PARP inhibition
Cell Death & Disease (2023)
-
Hypoxia-induced activation of NDR2 underlies brain metastases from Non-Small Cell Lung Cancer
Cell Death & Disease (2023)
-
E2 enzyme Bruce negatively regulates Hippo signaling through POSH-mediated expanded degradation
Cell Death & Disease (2023)