Abstract
Emerging evidence suggests that cancer is populated and maintained by tumour-initiating cells (TICs) with stem-like properties similar to those of adult tissue stem cells. Despite recent advances, the molecular regulatory mechanisms that may be shared between normal and malignant stem cells remain poorly understood. Here we show that the ΔNp63 isoform of the Trp63 transcription factor promotes normal mammary stem cell (MaSC) activity by increasing the expression of the Wnt receptor Fzd7, thereby enhancing Wnt signalling. Importantly, Fzd7-dependent enhancement of Wnt signalling by ΔNp63 also governs tumour-initiating activity of the basal subtype of breast cancer. These findings establish ΔNp63 as a key regulator of stem cells in both normal and malignant mammary tissues and provide direct evidence that breast cancer TICs and normal MaSCs share common regulatory mechanisms.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Visvader, J. E. Cells of origin in cancer. Nature 469, 314–322 (2011).
Nguyen, L. V., Vanner, R., Dirks, P. & Eaves, C. J. Cancer stem cells: an evolving concept. Nat. Rev. Cancer 12, 133–143 (2012).
Visvader, J. E. & Lindeman, G. J. Cancer stem cells: current status and evolving complexities. Cell Stem Cell 10, 717–728 (2012).
Magee, J. A., Piskounova, E. & Morrison, S. J. Cancer stem cells: impact, heterogeneity, and uncertainty. Cancer Cell 21, 283–296 (2012).
Lessard, J. & Sauvageau, G. Bmi-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature 423, 255–260 (2003).
Maddox, J. et al. Transcription factor Oct1 is a somatic and cancer stem cell determinant. PLoS Genet. 8, e1003048 (2012).
Cicalese, A. et al. The tumor suppressor p53 regulates polarity of self-renewing divisions in mammary stem cells. Cell 138, 1083–1095 (2009).
Guo, W. et al. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell 148, 1015–1028 (2012).
Kouros-Mehr, H. et al. GATA-3 links tumor differentiation and dissemination in a luminal breast cancer model. Cancer Cell 13, 141–152 (2008).
Chakrabarti, R. et al. Elf5 inhibits the epithelial-mesenchymal transition in mammary gland development and breast cancer metastasis by transcriptionally repressing Snail2. Nat. Cell Biol. 14, 1212–1222 (2012).
Crum, C. P. & McKeon, F. D. p63 in epithelial survival, germ cell surveillance, and neoplasia. Annu. Rev. Pathol. 5, 349–371 (2010).
Yang, A. et al. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol. Cell 2, 305–316 (1998).
Su, X., Chakravarti, D. & Flores, E. R. p63 steps into the limelight: crucial roles in the suppression of tumorigenesis and metastasis. Nat. Rev. Cancer 13, 136–143 (2013).
Deyoung, M. P. & Ellisen, L. W. p63 and p73 in human cancer: defining the network. Oncogene 26, 5169–5183 (2007).
Flores, E. R. et al. Tumor predisposition in mice mutant for p63 and p73: evidence for broader tumor suppressor functions for the p53 family. Cancer Cell 7, 363–373 (2005).
Keyes, W. M. et al. p63 heterozygous mutant mice are not prone to spontaneous or chemically induced tumors. Proc. Natl Acad. Sci. USA 103, 8435–8440 (2006).
Guo, X. et al. TAp63 induces senescence and suppresses tumorigenesis in vivo. Nat. Cell Biol. 11, 1451–1457 (2009).
Su, X. et al. TAp63 suppresses metastasis through coordinate regulation of Dicer and miRNAs. Nature 467, 986–990 (2010).
Karni-Schmidt, O. et al. Distinct expression profiles of p63 variants during urothelial development and bladder cancer progression. Am. J. Pathol. 178, 1350–1360 (2011).
Keyes, W. M. et al. DeltaNp63alpha is an oncogene that targets chromatin remodeler Lsh to drive skin stem cell proliferation and tumorigenesis. Cell Stem Cell 8, 164–176 (2011).
DiRenzo, J. et al. Growth factor requirements and basal phenotype of an immortalized mammary epithelial cell line. Cancer Res. 62, 89–98 (2002).
Teuliere, J. et al. Targeted activation of β-catenin signaling in basal mammary epithelial cells affects mammary development and leads to hyperplasia. Development 132, 267–277 (2005).
Li, N. et al. Reciprocal intraepithelial interactions between TP63 and hedgehog signaling regulate quiescence and activation of progenitor elaboration by mammary stem cells. Stem Cells 26, 1253–1264 (2008).
Regan, J. L. et al. c-Kit is required for growth and survival of the cells of origin of Brca1-mutation-associated breast cancer. Oncogene 31, 869–883 (2012).
Yalcin-Ozuysal, O. et al. Antagonistic roles of Notch and p63 in controlling mammary epithelial cell fates. Cell Death Differ. 17, 1600–1612 (2010).
Laurikkala, J. et al. p63 regulates multiple signalling pathways required for ectodermal organogenesis and differentiation. Development 133, 1553–1563 (2006).
Barton, C. E. et al. Novel p63 target genes involved in paracrine signaling and keratinocyte differentiation. Cell Death Dis. 1, e74 (2010).
Wu, N., Rollin, J., Masse, I., Lamartine, J. & Gidrol, X. p63 regulates human keratinocyte proliferation via MYC-regulated gene network and differentiation commitment through cell adhesion-related gene network. J. Biol. Chem. 287, 5627–5638 (2012).
Caserta, T. M. et al. p63 overexpression induces the expression of Sonic Hedgehog. Mol. Cancer Res. 4, 759–768 (2006).
Reya, T., Morrison, S. J., Clarke, M. F. & Weissman, I. L. Stem cells, cancer, and cancer stem cells. Nature 414, 105–111 (2001).
Takebe, N., Harris, P. J., Warren, R. Q. & Ivy, S. P. Targeting cancer stem cells by inhibiting Wnt, Notch, and Hedgehog pathways. Nat. Rev. Clin. Oncol. 8, 97–106 (2011).
Shackleton, M. et al. Generation of a functional mammary gland from a single stem cell. Nature 439, 84–88 (2006).
Van Amerongen, R., Bowman, A. N. & Nusse, R. Developmental stage and time dictate the fate of Wnt/β-catenin-responsive stem cells in the mammary gland. Cell Stem Cell 11, 387–400 (2012).
Zeng, Y. A. & Nusse, R. Wnt proteins are self-renewal factors for mammary stem cells and promote their long-term expansion in culture. Cell Stem Cell 6, 568–577 (2010).
Reya, T. & Clevers, H. Wnt signalling in stem cells and cancer. Nature 434, 843–850 (2005).
De La Coste, A. et al. Somatic mutations of the β-catenin gene are frequent in mouse and human hepatocellular carcinomas. Proc. Natl Acad. Sci. USA 95, 8847–8851 (1998).
Klaus, A. & Birchmeier, W. Wnt signalling and its impact on development and cancer. Nat. Rev. Cancer 8, 387–398 (2008).
Smid, M. et al. Subtypes of breast cancer show preferential site of relapse. Cancer Res. 68, 3108–3114 (2008).
Khramtsov, A. I. et al. Wnt/β-catenin pathway activation is enriched in basal-like breast cancers and predicts poor outcome. Am. J. Pathol. 176, 2911–2920 (2010).
Howe, L. R. & Brown, A. M. Wnt signaling and breast cancer. Cancer Biol. Ther. 3, 36–41 (2004).
Alexander, C. M., Goel, S., Fakhraldeen, S. A. & Kim, S. Wnt signaling in mammary glands: plastic cell fates and combinatorial signaling. Cold Spring Harb. Perspect. Biol. 4, a008037 (2012).
Vaillant, F. et al. The mammary progenitor marker CD61/β3 integrin identifies cancer stem cells in mouse models of mammary tumorigenesis. Cancer Res. 68, 7711–7717 (2008).
Romano, R. A. et al. DeltaNp63 knockout mice reveal its indispensable role as a master regulator of epithelial development and differentiation. Development 139, 772–782 (2012).
Kouwenhoven, E. N. et al. Genome-wide profiling of p63 DNA-binding sites identifies an element that regulates gene expression during limb development in the 7q21 SHFM1 locus. PLoS Genet. 6, e1001065 (2010).
Karantza-Wadsworth, V. & White, E. A mouse mammary epithelial cell model to identify molecular mechanisms regulating breast cancer progression. Methods Enzymol. 446, 61–76 (2008).
Cancer Genome Atlas, Network Comprehensive molecular portraits of human breast tumours. Nature 490, 61–70 (2012).
Wang, Y. et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet 365, 671–679 (2005).
DeRose, Y. S. et al. Tumor grafts derived from women with breast cancer authentically reflect tumor pathology, growth, metastasis and disease outcomes. Nat. Med. 17, 1514–1520 (2011).
Herschkowitz, J. I. et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 8, R76 (2007).
Lim, E. et al. Transcriptome analyses of mouse and human mammary cell subpopulations reveal multiple conserved genes and pathways. Breast Cancer Res. 12, R21 (2010).
Cho, R. W. et al. Isolation and molecular characterization of cancer stem cells in MMTV-Wnt-1 murine breast tumors. Stem Cells 26, 364–371 (2008).
Carroll, D. K. et al. p63 regulates an adhesion programme and cell survival in epithelial cells. Nat. Cell Biol. 8, 551–561 (2006).
Makarem, M. et al. Developmental changes in the in vitro activated regenerative activity of primitive mammary epithelial cells. PLoS Biol. 11, e1001630 (2013).
Balboni, A. L. et al. ΔNp63α-mediated activation of bone morphogenetic protein signaling governs stem cell activity and plasticity in normal and malignant mammary epithelial cells. Cancer Res. 73, 1020–1030 (2013).
Metzger-Filho, O. et al. Dissecting the heterogeneity of triple-negative breast cancer. J. Clin. Oncol. 30, 1879–1887 (2012).
Lehmann, B. D. et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J. Clin. Invest. 121, 2750–2767 (2011).
Yang, L. et al. FZD7 has a critical role in cell proliferation in triple negative breast cancer. Oncogene 30, 4437–4446 (2011).
King, T. D., Zhang, W., Suto, M. J. & Li, Y. Frizzled7 as an emerging target for cancer therapy. Cell. Signal. 24, 846–851 (2012).
Kinzler, K. W. & Vogelstein, B. Lessons from hereditary colorectal cancer. Cell 87, 159–170 (1996).
Molyneux, G. & Smalley, M. J. The cell of origin of BRCA1 mutation-associated breast cancer: a cautionary tale of gene expression profiling. J. Mammary Gland Biol. Neoplasia 16, 51–55 (2011).
Lim, E. et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat. Med. 15, 907–913 (2009).
Eirew, P. et al. A method for quantifying normal human mammary epithelial stem cells with in vivo regenerative ability. Nat. Med. 14, 1384–1389 (2008).
Asselin-Labat, M. L. et al. Control of mammary stem cell function by steroid hormone signalling. Nature 465, 798–802 (2010).
Kannan, N. et al. Glutathione-dependent and -independent oxidative stress-control mechanisms distinguish normal human mammary epithelial cell subsets. Proc. Natl Acad. Sci. USA 110, 7789–7794 (2014).
Matveeva, O. V. et al. Optimization of duplex stability and terminal asymmetry for shRNA design. PloS ONE 5, e10180 (2010).
Brummelkamp, T. R., Bernards, R. & Agami, R. Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer Cell 2, 243–247 (2002).
Fuerer, C. & Nusse, R. Lentiviral vectors to probe and manipulate the Wnt signaling pathway. PloS ONE 5, e9370 (2010).
Acknowledgements
We thank our laboratory members for helpful discussions, R. A. Weinberg and V. Karantza for iMMEC and HMLE cell lines, and A. L. Welm and M. T. Lewis for PDX lines, respectively. This research was supported by grants from the Brewster Foundation, the Department of Defense (BC123187) and the National Institutes of Health (R01CA134519 and R01CA141062) to Y.K. and from the Canadian Cancer Society Research Institute to C.J.E. R.C. and N.K. are recipients of postdoctoral fellowships from the Department of Defense and MITACS, respectively. This research was also supported by the Transgenic/Knockout, Tissue Analytic Service and Flow Cytometry Shared Resources of the Cancer Institute of New Jersey (P30CA072720).
Author information
Authors and Affiliations
Contributions
R.C. and Y.K. designed experiments. R.C., Y.W., J.H., X.H., M.A.B., A.C., B.T., N.K. and S.S. performed the experiments. T.I., L.M., D.A., R-A.R., C.D., N.K. and C.J.E. provided crucial samples and technical advice. R.C. and Y.K. wrote the manuscript. All authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 ΔNp63 is enriched in MaSCs and promotes mammary stem cell activity.
(a) Oocyte in mouse ovary sections stained with TAp63 antibody as positive control for the antibody. (b) Phase contrast and fluorescence images of GFP-expressing MECs 72 h post-lentiviral transduction. (c) Whole mounts of GFP+ ductal outgrowths derived from P4 cells transduced with the GFP control vector. (d,e) Overexpression of ΔNp63 and TAp63 in primary mammary epithelial cells (MECs) transduced with ΔNp63 and TAp63 compared to control empty vector, as demonstrated by qRT-PCR (n = 3 samples; data represents mean ± s.d.) (d) and western blot (e). qRT-PCR values were normalized to the housekeeping gene Gapdh. ∗p < 0.05 by Student’s t test. (f) P4 population were transduced with control or ΔNp63 expressing lentiviruses and were transplanted into cleared fat pads and take rate was compared after using limiting numbers of cells in injections (n = number of mammary fat pad injections as indicated in the table). P value was obtained by Pearson’s χ2 test using ELDA software. (g) Percentage of fat pad filled from experiments in f. (h) P5 population were transduced with control or ΔNp63 expressing lentiviruses and were transplanted into cleared fat pads and take rate was compared after using limiting numbers of cells in injections (n = number of mammary fat pad injections as indicated in the table). P value was obtained by Pearson’s χ2 test using ELDA software. (i) Percentage of fat pad filled from experiments in h. (j) Competitive reconstitution assay strategy for MaSCs (2000 cells per injection) after lentiviral transduction of ΔNp63, TAp63 or control expression plasmids. Size bars, 75 μm in a, and 40 μm in b and c respectively. Uncropped images of blots are shown in Supplementary Fig. 9.
Supplementary Figure 2 ΔNp63 increases mammary stem cell-like functions in P5-CD61+ and P5-CD61− luminal cells but not in the P6 population.
(a) Table showing reconstitution efficiency of limiting dilution of CD61+ or CD61− cells from Lin−CD24+CD29lo (P5) cells with or without ΔNp63 overexpression (n = number of mammary fat pad injections as indicated in the table). (b) Representative alum carmine stained mammary outgrowths from (a). (c) Representative images of 3-D matrigel colonies formed by control and ΔNp63 overexpressing P5 cells (upper panels) as well as K14 and K8 staining of colonies (lower panels). (d) K14 and K8 staining in mammary outgrowths from control and ΔNp63 overexpressing CD61+ or CD61− cells from Lin−CD24+CD29lo (P5) population. (e) Table showing reconstitution efficiency of limiting dilution of P6 cells with or without ΔNp63 or TAp63 (n = number of mammary fat pad injections as indicated in the table). (f) Representative alum carmine stained mammary outgrowths from (e). Size bar,1 mm in b,f and 40 μm in c and d. p value was obtained by Pearson’s χ2 test using ELDA software in a and e.
Supplementary Figure 3 ΔNp63 increases proliferation and blocks differentiation in the mammary epithelium.
(a) H&E stained outgrowth sections from control and ΔNp63 overexpressing cells. Quantification of ductal branching (tertiary branch points in defined area) is shown at the bottom panel (n = 10 areas scored for branch points from 3 mammary outgrowths per group). p-value computed by Mann-Whitney U test. Box represents 75th, 50th and 25th percentile of the values. The top and bottom lines represent the maximum and minimal data points within 1.5 × IQ (inter quarter) range, respectively. (b) Ki67 staining in outgrowth sections from control and ΔNp63 overexpressing cells. Quantification of Ki67+ cell percentage over total epithelial cells in field of view is shown in the bottom panel (n = 25 areas scored from 3 mammary outgrowths per group, data represents mean ± s.d.). (c) ΔNp63 staining in outgrowth sections from control (n = 10 ducts and TEB sections scored from 3 mammary outgrowths; data represents mean ± s.d.) and ΔNp63 overexpressing (n = 12 ducts and TEB sections scored from 3 mammary outgrowths; data represents mean ± s.d.) cells. Quantification of the percentage ΔNp63+ basal cells among all basal cells was shown in the bottom panel. (d) K14 and K8 staining in mammary outgrowths from control (n = 10 ducts and TEB sections scored from 3 mammary outgrowths; data represents mean ± s.d.) and ΔNp63 overexpressing (n = 12 ducts and TEB sections scored from 3 mammary outgrowths; data represents mean ± s.d.) cells. Quantification of the number of K14+ basal cells in the field of view is shown in the bottom panel. P value computed by Student’s t test. (e) Western blot analyses of ΔNp63 and K14 in mammary epithelial cells from control and ΔNp63 overexpressing outgrowths. (f,g) Representative FACS plot of mammary epithelial cells from control and ΔNp63 overexpressing outgrowths after staining with CD24 and CD29. Results were quantified in g. P value computed by paired t test. Size bar, 100 μm in a, 40 μm in b,c and d. Uncropped images of blots are shown in Supplementary Fig. 9.
Supplementary Figure 4 ΔNp63 regulation of MaSC activity is dependent on its DNA-binding activity.
(a,b) Representative images (a) and quantification (b) of 3-D matrigel colonies (1st generation) formed by control, ΔNp63 and TAp63 overexpressing P4 cells (n = 5 samples; data represents mean ± s.d.). Colony images from lower panel are representative single colonies in higher magnifications from upper panel images were in a. (c) Representative image of outgrowths from 3rd generation shown in Fig. 1k. (d,e) qRT-PCR analysis (n = 6 technical replicates pooled from 2 independent experiments; data represents mean ± s.d.) (d) and western blot analyses (e) of the expression of ΔNp63 in control, WT ΔNp63 (replicate populations) and DNA binding mutant form of ΔNp63 overexpressing MECs 72 hr post-transduction. qRT-PCR values were normalized to the housekeeping gene Gapdh. (f,g) Bar graphs (f) and representative images (g) of colonies formed in 3-D matrigel colony formation assay from control, WT and DNA binding mutant of ΔNp63 overexpressing Lin−CD24+CD29hi (P4) cells (n = 4 samples; data represents mean ± s.d.) in f. (h) P4 cells were transduced with control, WT, or mutant ΔNp63 expressing lentiviruses were transplanted into cleared fat pads and take rate was compared (n = number of mammary fat pad injections as indicated in the table). ∗p < 0.05 by Student’s t test. P value was obtained by Pearson’s χ2 test using ELDA software in h. Size bar, 1 mm (upper panel) and 40 μm (lower panel) in a, 2 mm in c and 1 mm in g, respectively. Uncropped images of blots are shown in Supplementary Fig. 9.
Supplementary Figure 5 ΔNp63gfp/+ mice exhibit defects in ductal morphogenesis and MaSCs defect at 9-10 weeks.
(a) Schematic diagram of generation of ΔNp63gfp/+ mice. Targeting vector showing the insertion of a GFP coding cassette in-frame with the start codon of ΔNp63 located in exon 3′, followed by transcription termination signal (PolyA), leading to loss of expression of the endogenous ΔNp63. (b) Quantification of ductal elongation (arbitrary units) (left) (n = 5 mammary glands per group) and percentage (right) of MaSC-enriched (P4) cells in WT and ΔNp63gfp/+ mammary glands (6-7 weeks). For the right panel, WT (n = 8 mammary glands) and ΔNp63gfp/+ mice (n = 12 mammary glands). p-value computed by Mann-Whitney U test. Box represents 75th, 50th and 25th percentile of the values. The top and bottom lines represent the maximum and minimal data points within 1.5 × IQ (inter quarter) range, respectively. (c) Representative alum carmine stained whole mount mammary outgrowths from WT and ΔNp63gfp/+ mice at 9-10 weeks. (d) Quantification of ductal elongation (arbitrary units) (left) and percentage (right) of MaSC-enriched (P4) cells in WT and ΔNp63gfp/+ mammary glands (9-10 weeks) (n = 5 mammary glands per group). p-value computed by Mann-Whitney U test. Box represents 75th, 50th and 25th percentile of the values. The top and bottom lines represent the maximum and minimal data points within 1.5 × IQ (inter quarter) range, respectively. (e) Representative FACS profiles of mammary epithelial cells from WT and ΔNp63gfp/+ mice after dissociation and staining with CD24 and CD29. (f) Bar graph showing the number of colonies formed in Matrigel from control or ΔNp63 overexpressing P4 cells (20,000 cells) treated with Dikkopf-1 (Dkk1) for 7 days (n = 3 samples; data represents mean ± s.d.). (g) Bar graph showing colonies from 1st generation that were dissociated and passaged with equal cell numbers for 3 generations with Dkk1 treatment (n = 3 samples; data represents mean ± s.d.). ∗p < 0.05 by Student’s t test in f and g(h) Representative images of 3-D matrigel colonies from 3rd generation P4 cells with or without ΔNp63 overexpression. Size bar, 1 mm (left panel), 2 mm (right panel) in c, respectively and 1 mm in h.
Supplementary Figure 6 Wnt signalling is necessary for ΔNp63-induced mammary stem cell activity.
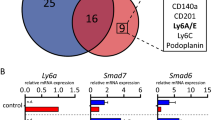
(a) Venn diagram showing significant overlap of core genes from two Wnt signalling GSEA signatures. Common core genes between 2 sets are presented in the table. (b) qRT-PCR analyses of the expression of ΔNp63 and TAp63 in control, ΔNp63, or TAp63 overexpressing HMLE cells (n = 3 samples; data represents mean ± s.d.).∗p < 0.05 by Student’s t test. (c,d) Representative alum carmine stained mammary outgrowth images (c) and quantification table (d) showing reconstitution efficiency of P4 cells of indicated groups (n = number of mammary fat pad injections as indicated in the table). P value was obtained by Pearson’s χ2 test using ELDA software. (e) GSEA data showing positive enrichment of published WNT gene signatures in ΔNp63 signaturehi patients compared to ΔNp63 signaturelo patients. Patients were stratified using an experimentally-derived ΔNp63-specific gene expression signature as described in methods section. (f) Scatter plot showing the correlation between ΔNp63signature scores and FZD7 expression in clinical patients from EMC286 dataset (n = 286 patients). Pearson Coefficient test was performed for statistical significance. (g) Percentage of tumours with positive detection of ΔNp63 and TAp63expression from the TCGA-BRCA-RNAseqV2 clinical dataset (n = 1000 patients). Size bar, 1 mm in c.
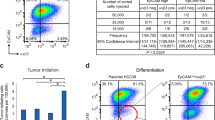
Supplementary Figure 7 ΔNp63 and Fzd7 expression and function in human and mouse mammary tumours.
(a) Western blot analysis of ΔNp63 and FZD7 expression in MDA-MB-231 cells transduced with shRNAs targeting p63 or FZD7, or control shRNA. (b,c) Tumor incidence of MDA-MB-231 (b) and SUM-1315 cells (c) with or without p63 KD. Tumor incidence was determined 70 days post injection (n = number of mammary fat pad injections as indicated in the table). p value was obtained by Pearson’s χ2 test using ELDA software. (d) Confocal images of immunofluorescence analysis of ΔNp63, TAp63 and Fzd7 protein expression in human patient derived xenografts (PDX). (e) Immunostaining of protein expression showing predominant expression of ΔNp63 and Fzd7 in basal (MMTV-Wnt1) compared to luminal (MMTV-PyMT) type mammary gland tumours. Mixed (MMTV-cMyc) type has modest expression of ΔNp63 and Fzd7. (f) qRT-PCR analysis of the RNA expression of ΔNp63 and TAp63 in MMTV-Wnt-1 tumor cells (n = 3 tumors; data represents mean ± s.d.). qRT-PCR values were normalized to the housekeeping gene Gapdh. ∗p < 0.05 by Student’s t test. Size bar, 40 μm in d and 100 μm in e respectively. Uncropped images of blots are shown in Supplementary Fig. 9.
Supplementary Figure 8 Reduced Fzd7 expression in MMTV-Wnt1; ΔNp63gfp/+tumors compared to MMTV-Wnt1; ΔNp63+/+ tumours.
(a) qRT-PCR analysis of ΔNp63, Fzd7 and Wnt5a in the in MMTV-Wnt1; ΔNp63+/+ and MMTV-Wnt1; ΔNp63gfp/+ tumours (n = 5 tumours per genotype). ∗p < 0.05 by Mann Whitney U test. (b) Western blot analyses of indicated protein expression in MMTV-Wnt1; ΔNp63+/+ and MMTV-Wnt1; ΔNp63gfp/+ tumours. (c) Confocal immunofluorescence images of ΔNp63, K14 and K8 and Fzd7 protein expression in MMTV-Wnt1; ΔNp63+/+ and MMTV-Wnt1; ΔNp63gfp/+ tumours. Size bar, 40 μm in c. Uncropped images of blots are shown in Supplementary Fig. 9.
Supplementary information
Supplementary Information
Supplementary Information (PDF 3509 kb)
Rights and permissions
About this article
Cite this article
Chakrabarti, R., Wei, Y., Hwang, J. et al. ΔNp63 promotes stem cell activity in mammary gland development and basal-like breast cancer by enhancing Fzd7 expression and Wnt signalling. Nat Cell Biol 16, 1004–1015 (2014). https://doi.org/10.1038/ncb3040
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb3040
This article is cited by
-
Mcam inhibits macrophage-mediated development of mammary gland through non-canonical Wnt signaling
Nature Communications (2024)
-
Reactivation of embryonic genetic programs in tissue regeneration and disease
Nature Genetics (2023)
-
p63: a crucial player in epithelial stemness regulation
Oncogene (2023)
-
The implications of alternative pre-mRNA splicing in cell signal transduction
Experimental & Molecular Medicine (2023)
-
FZD7 accelerates hepatic metastases in pancreatic cancer by strengthening EMT and stemness associated with TGF-β/SMAD3 signaling
Molecular Medicine (2022)