Abstract
Although aberrant reactivation of embryonic gene programs is intricately linked to pathological heart disease, the transcription factors driving these gene programs remain ill-defined. Here we report that increased calcineurin/Nfat signalling and decreased miR-25 expression integrate to re-express the basic helix-loop-helix (bHLH) transcription factor dHAND (also known as Hand2) in the diseased human and mouse myocardium. In line, mutant mice overexpressing Hand2 in otherwise healthy heart muscle cells developed a phenotype of pathological hypertrophy. Conversely, conditional gene-targeted Hand2 mice demonstrated a marked resistance to pressure-overload-induced hypertrophy, fibrosis, ventricular dysfunction and induction of a fetal gene program. Furthermore, in vivo inhibition of miR-25 by a specific antagomir evoked spontaneous cardiac dysfunction and sensitized the murine myocardium to heart failure in a Hand2-dependent manner. Our results reveal that signalling cascades integrate with microRNAs to induce the expression of the bHLH transcription factor Hand2 in the postnatal mammalian myocardium with impact on embryonic gene programs in heart failure.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Accession codes
References
Olson, E. N. & Schneider, M. D. Sizing up the heart: development redux in disease. Genes Dev. 17, 1937–1956 (2003).
Katz, A. M. The cardiomyopathy of overload: an unnatural growth response. Eur. Heart J. 16, 110–114 (1995).
Hoshijima, M. & Chien, K. R. Mixed signals in heart failure: cancer rules. J. Clin. Invest. 109, 849–855 (2002).
Eulalio, A. et al. Functional screening identifies miRNAs inducing cardiac regeneration. Nature 492, 376–381 (2012).
Senyo, S. E. et al. Mammalian heart renewal by pre-existing cardiomyocytes. Nature 493, 433–436 (2013).
Towbin, J. A. & Bowles, N. E. The failing heart. Nature 415, 227–233 (2002).
Oka, T., Xu, J. & Molkentin, J. D. Re-employment of developmental transcription factors in adult heart disease. Semin. Cell Dev. Biol. 18, 117–131 (2007).
Heineke, J. & Molkentin, J. D. Regulation of cardiac hypertrophy by intracellular signalling pathways. Nat. Rev. Mol. Cell Biol. 7, 589–600 (2006).
Savage, D. D., Levy, D., Dannenberg, A. L., Garrison, R. J. & Castelli, W. P. Association of echocardiographic left ventricular mass with body size, blood pressure and physical activity (the Framingham Study). Am. J. Cardiol. 65, 371–376 (1990).
Meijs, M. F. et al. Left ventricular hypertrophy: a shift in paradigm. Curr. Med. Chem. 14, 157–171 (2007).
Aries, A., Paradis, P., Lefebvre, C., Schwartz, R. J. & Nemer, M. Essential role of GATA-4 in cell survival and drug-induced cardiotoxicity. Proc. Natl Acad. Sci. USA 101, 6975–6980 (2004).
Oka, T. et al. Cardiac-specific deletion of Gata4 reveals its requirement for hypertrophy, compensation, and myocyte viability. Circ. Res. 98, 837–845 (2006).
Bar, H., Kreuzer, J., Cojoc, A. & Jahn, L. Upregulation of embryonic transcription factors in right ventricular hypertrophy. Basic Res. Cardiol. 98, 285–294 (2003).
Song, K. et al. The transcriptional coactivator CAMTA2 stimulates cardiac growth by opposing class II histone deacetylases. Cell 125, 453–466 (2006).
Bourajjaj, M. et al. NFATc2 is a necessary mediator of calcineurin-dependent cardiac hypertrophy and heart failure. J. Biol. Chem. 283, 22295–22303 (2008).
Molkentin, J. D. et al. A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell 93, 215–228 (1998).
Freund, C. et al. Requirement of nuclear factor-κB in angiotensin II- and isoproterenol-induced cardiac hypertrophy in vivo. Circulation 111, 2319–2325 (2005).
Li, Y. et al. NF-κB activation is required for the development of cardiac hypertrophy in vivo. Am. J. Physiol. Heart Circ. Physiol. 287, H1712–H1720 (2004).
Wang, J. et al. Targeted disruption of Smad4 in cardiomyocytes results in cardiac hypertrophy and heart failure. Circ. Res. 97, 821–828 (2005).
Xu, J. et al. GDF15/MIC-1 functions as a protective and antihypertrophic factor released from the myocardium in association with SMAD protein activation. Circ. Res. 98, 342–350 (2006).
Buitrago, M. et al. The transcriptional repressor Nab1 is a specific regulator of pathological cardiac hypertrophy. Nat. Med. 11, 837–844 (2005).
Saadane, N., Alpert, L. & Chalifour, L. E. Altered molecular response to adrenoreceptor-induced cardiac hypertrophy in Egr-1-deficient mice. Am. J. Physiol. Heart Circ. Physiol. 278, H796–H805 (2000).
Srivastava, D., Cserjesi, P. & Olson, E. N. A subclass of bHLH proteins required for cardiac morphogenesis. Science 270, 1995–1999 (1995).
Cross, J. C. et al. Hxt encodes a basic helix-loop-helix transcription factor that regulates trophoblast cell development. Development 121, 2513–2523 (1995).
Hollenberg, S. M., Sternglanz, R., Cheng, P. F. & Weintraub, H. Identification of a new family of tissue-specific basic helix-loop-helix proteins with a two-hybrid system. Mol. Cell Biol. 15, 3813–3822 (1995).
Srivastava, D. et al. Regulation of cardiac mesodermal and neural crest development by the bHLH transcription factor, dHAND. Nature Genet. 16, 154–160 (1997).
Biben, C. & Harvey, R. P. Homeodomain factor Nkx2-5 controls left/right asymmetric expression of bHLH gene eHand during murine heart development. Genes Dev. 11, 1357–1369 (1997).
Thomas, T., Yamagishi, H., Overbeek, P. A., Olson, E. N. & Srivastava, D. The bHLH factors, dHAND and eHAND, specify pulmonary and systemic cardiac ventricles independent of left-right sidedness. Dev. Biol. 196, 228–236 (1998).
Morikawa, Y. & Cserjesi, P. Cardiac neural crest expression of Hand2 regulates outflow and second heart field development. Circ. Res. 103, 1422–1429 (2008).
Snider, P., Olaopa, M., Firulli, A. B. & Conway, S. J. Cardiovascular development and the colonizing cardiac neural crest lineage. Sci. World J. 7, 1090–1113 (2007).
Hutson, M. R. & Kirby, M. L. Model systems for the study of heart development and disease. Cardiac neural crest and conotruncal malformations. Semin. Cell Dev. Biol. 18, 101–110 (2007).
Rockman, H. A. et al. Segregation of atrial-specific and inducible expression of an atrial natriuretic factor transgene in an in vivo murine model of cardiac hypertrophy. Proc. Natl Acad. Sci. USA 88, 8277–8281 (1991).
Subramaniam, A. et al. Tissue-specific regulation of the alpha-myosin heavy chain gene promoter in transgenic mice. J. Biol. Chem. 266, 24613–24620 (1991).
De Windt, L. J. et al. Calcineurin-mediated hypertrophy protects cardiomyocytes from apoptosis in vitro and in vivo: an apoptosis-independent model of dilated heart failure. Circ. Res. 86, 255–263 (2000).
Zhang, Z. et al. The microRNA-processing enzyme Dicer is dispensable for somite segmentation but essential for limb bud positioning. Dev. Biol. 351, 254–265 (2011).
Sohal, D. S. et al. Temporally regulated and tissue-specific gene manipulations in the adult and embryonic heart using a tamoxifen-inducible Cre protein. Circ. Res. 89, 20–25 (2001).
Koitabashi, N. et al. Avoidance of transient cardiomyopathy in cardiomyocyte-targeted tamoxifen-induced MerCreMer gene deletion models. Circ. Res. 105, 12–15 (2009).
Koitabashi, N. et al. Pivotal role of cardiomyocyte TGF-beta signaling in the murine pathological response to sustained pressure overload. J. Clin. Invest. 121, 2301–2312 (2011).
Fish, J. E. et al. A Slit/miR-218/Robo regulatory loop is required during heart tube formation in zebrafish. Development 138, 1409–1419 (2011).
Baker, R. K., Vanderboom, A. K., Bell, G. W. & Antin, P. B. Expression of the receptor tyrosine kinase gene EphB3 during early stages of chick embryo development. Mech. Dev. 104, 129–132 (2001).
Mason, I. J., Fuller-Pace, F., Smith, R. & Dickson, C. FGF-7 (keratinocyte growth factor) expression during mouse development suggests roles in myogenesis, forebrain regionalisation and epithelial-mesenchymal interactions. Mech. Dev. 45, 15–30 (1994).
Dai, Y. S. & Cserjesi, P. The basic helix-loop-helix factor, HAND2, functions as a transcriptional activator by binding to E-boxes as a heterodimer. J. Biol. Chem. 277, 12604–12612 (2002).
McFadden, D. G. et al. A GATA-dependent right ventricular enhancer controls dHAND transcription in the developing heart. Development 127, 5331–5341 (2000).
Zhao, Y., Samal, E. & Srivastava, D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature 436, 214–220 (2005).
Tanzer, A. & Stadler, P. F. Molecular evolution of a microRNA cluster. J. Mol. Biol. 339, 327–335 (2004).
Aramburu, J. et al. Affinity-driven peptide selection of an NFAT inhibitor more selective than cyclosporin A. Science 285, 2129–2133 (1999).
Care, A. et al. MicroRNA-133 controls cardiac hypertrophy. Nat. Med. 13, 613–618 (2007).
da Costa Martins, P. A. et al. MicroRNA-199b targets the nuclear kinase Dyrk1a in an auto-amplification loop promoting calcineurin/NFAT signalling. Nature Cell Biol. 12, 1220–1227 (2010).
Ganesan, J. et al. MiR-378 controls cardiac hypertrophy by combined repression of mitogen-activated protein kinase pathway factors. Circulation 127, 2097–2106 (2013).
Van Rooij, E. et al. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science 316, 575–579 (2007).
Ucar, A. et al. The miRNA-212/132 family regulates both cardiac hypertrophy and cardiomyocyte autophagy. Nat. Commun. 3, 1078 (2012).
Boon, R. A. et al. MicroRNA-34a regulates cardiac ageing and function. Nature 495, 107–110 (2013).
Thum, T. et al. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature 456, 980–984 (2008).
Bonauer, A. et al. MicroRNA-92a controls angiogenesis and functional recovery of ischemic tissues in mice. Science 324, 1710–1713 (2009).
Morikawa, Y., D’Autreaux, F., Gershon, M. D. & Cserjesi, P. Hand2 determines the noradrenergic phenotype in the mouse sympathetic nervous system. Dev. Biol. 307, 114–126 (2007).
Krutzfeldt, J. et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature 438, 685–689 (2005).
Lyakh, L., Ghosh, P. & Rice, N. R. Expression of NFAT-family proteins in normal human T cells. Mol. Cell Biol. 17, 2475–2484 (1997).
Wilkinson, D. G., Bailes, J. A., Champion, J. E. & McMahon, A. P. A molecular analysis of mouse development from 8 to 10 days post coitum detects changes only in embryonic globin expression. Development 99, 493–500 (1987).
De Windt, L. J., Lim, H. W., Haq, S., Force, T. & Molkentin, J. D. Calcineurin promotes protein kinase C and c-Jun NH2-terminal kinase activation in the heart. Cross-talk between cardiac hypertrophic signaling pathways. J. Biol. Chem. 275, 13571–13579 (2000).
Van Oort, R. J. et al. MEF2 activates a genetic program promoting chamber dilation and contractile dysfunction in calcineurin-induced heart failure. Circulation 114, 298–308 (2006).
Liang, Q. et al. The transcription factors GATA4 and GATA6 regulate cardiomyocyte hypertrophy in vitro and in vivo. J. Biol. Chem. 276, 30245–30253 (2001).
Van Rooij, E. et al. Requirement of nuclear factor of activated T-cells incalcineurin-mediated cardiomyocyte hypertrophy. J. Biol. Chem. 277, 48617–48626 (2002).
Armand, A. S. et al. Cooperative synergy between NFAT and MyoDregulates myogenin expression and myogenesis. J. Biol. Chem. 283, 29004–29010 (2008).
Luo, C. et al. Recombinant NFAT1 (NFATp) is regulated by calcineurin in T cells and mediates transcription of several cytokine genes. Mol. Cell Biol. 16, 3955–3966 (1996).
Acknowledgements
We gratefully acknowledge G. Summer for bioinformatics help and G. van Hout for technical support. T.T. was supported by the German Research Foundation (TH903/11-1) and the REBIRTH Exellence Cluster. T.E. was supported by the German Research Foundation (ES88/12-1) and the DZHK, the German Centre for Cardiovascular Research funded by the German Ministry of Research and Education (BMBF). P.A.d.C.M. was supported by a Leducq Career Development Award and the Dutch Heart Foundation grant NHS2010B261. L.J.D.W. acknowledges support from the Netherlands CardioVascular Research Initiative: the Dutch Heart Foundation, Dutch Federation of University Medical Centers, the Netherlands Organization for Health Research and Development (ZonMW) and the Royal Netherlands Academy of Sciences. L.J.D.W. was further supported by a VIDI award 917-863-72 from the ZonMW; the Dutch Heart Foundation program grant NHS2007B167; the Fondation Leducq Transatlantic Network of Excellence program 08-CVD-03 and grant 311549 from the European Research Council (ERC).
Author information
Authors and Affiliations
Contributions
E.D., P.A.d.C.M, M.M.G., L.E.P., K.S., M.B. and N.K. performed northern blots and quantitative real-time PCR experiments. M.M.G., S.O. and S.S. performed western blots. E.D. and M.M.G. performed luciferase assays. S.L. performed bioinformatics analyses. E.D., V.K. and S.O. performed chromatin immunoprecipitation assays. A-S.A. and C.C. performed mouse embryo studies. Y.M., P.C., S.O. and L.J.D.W. created genetically modified mouse models. H.e.A. and N.B. performed surgical procedures in mouse models. E.D., P.A.d.C.M., M.M.G., R.N., K.S., N.B. and G.J.J.d.S. performed echocardiography and histology in mouse models. E.D., P.A.d.C.M., M.M.G., A-S.A., K.S., S.L. and L.J.D.W. analysed data. C.C., S.H., P.G.A.V., T.T., S.D., P.C. and T.E. provided reagents, models or data. E.D., P.A.d.C.M. and L.J.D.W. designed the study. E.D., P.A.d.C.M. and L.J.D.W. wrote the manuscript. P.A.d.C.M. and L.J.D.W. acquired funding for the study. E.D. and M.M.G. contributed equally as joint first authors. P.A.d.C.M. and L.J.D.W. contributed equally as joint last authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
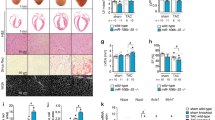
Supplementary Figure 1 Additive cardiomyocyte hypertrophy by Hand2 overexpression in combination with overexpression with MEF2, NFAT or GATA4.
(a) Confocal microscopy images of neonatal rat cardiomyocytes infected with AdGFP, AdMEF2A, AdNFAT or AdGATA4 alone (top row). Confocal microscopy images of neonatal rat cardiomyocyte pre-infected with AdHand2 and infected with AdGFP, AdMEF2A, AdNFAT or AdGATA4 (bottom row). Nuclei were visualized with DAPI and cells were stained with an antibody against α-actinin (red). Scale bar, 50 μm. (b) Quantification of cell surface area in conditions in (a), n refers to number of microscopic fields. *P<0.05 vs control group; #P<0.05 vs experimental group (error bars are s.e.m.). Source data are shown in Supplementary Table 9.
Supplementary Figure 2 Effect of tamoxifen treatment on cardiac function.
(a) Real-time PCR analysis of Hand2, Nppa and Nppb transcript abundance in hearts from MHC-MerCreMer transgenic (MHC-MCM) mice treated with vehicle or tamoxifen at a dose of 40 mg/kg/day for 5 consecutive days dissolved in sunflower oil as described previously or tamoxifen at a dose of 45 mg/kg/day for 5 consecutive days dissolved in peanut oil (this study), n refers to number of hearts. (b) Quantification of the left ventricular mass, (c) fractional shortening (FS), (d) left ventricular internal diameter in systole (LVIDs) and (e) E/A ratio from pulsed-wave Doppler imaging of the ratio of blood flow through the mitral valve during early (E) versus late (A) diastole in MHC-MCM mice, injected with vehicle (PBS) or 45 mg/kg/day tamoxifen dissolved in peanut oil, n refers to number of animals. (f) Real-time PCR analysis of Rcan1.4 expression in hearts from mice with indicated genotype, pre-treated with vehicle or tamoxifen and subjected to sham or TAC surgery, n refers to number of hearts. *P<0.05 vs control group; #P<0.05 vs experimental group (error bars are s.e.m.). Source data are shown in Supplementary Table 9.
Supplementary Figure 3 Real-time PCR validation of microarray results in pressure overloaded Hand2F/F and MCM-Hand2F/F mice.
(a) Validation of the gene profiling microarray results from Supplementary Tables 1 and 2 by quantitative real-time PCR, n refers to number of hearts. Relative mRNA expression levels were determined for transcripts that were increased in expression in pressure overloaded Hand2-deficient hearts (Abcc8, Ephb3, Smad6) or (b) downregulated in pressure overloaded Hand2-deficient hearts (Enpp1, Cacna1c, Lama2, Actb, Robo2 and Id1), n refers to number of hearts. *P<0.05 vs control group (error bars are s.e.m.). Source data are shown in Supplementary Table 9.
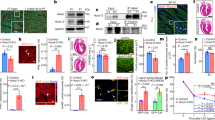
Supplementary Figure 4 miR-25, miR-92a, miR-92b and miR-1 expression levels in heart disease.
(a) Location of potential miR-92a, miR-92b and miR-1 seed regions in human and mouse Hand2 3′UTR. (b) Northern blot analysis of miR-92a expression in hearts from nontransgenic littermates (nTg), calcineurin transgenic mice (MHC-CnA) and mice subjected to transverse aortic constriction (TAC). Quantification of Rnu6-2 corrected Northern blot signals for miR-92a (right panel), n refers to number of hearts. (c) Northern blot analysis of miR-92b expression in hearts from nontransgenic littermates (nTg), calcineurin transgenic mice (MHC-CnA) and mice subjected to transverse aortic constriction (TAC). Quantification of Rnu6-2 corrected northern blot miR-92b signals (right panel), n refers to number of hearts. (d) Northern blot analysis of miR-1 expression levels in hearts from nTg mice, MHC-CnA mice and mice subjected to TAC. Quantification of Rnu6-2 corrected Northern blot miR-1 signals (right panel), n refers to number of hearts. (e) Real-time PCR analysis of miR-92a expression in hearts from mice treated with scrambled antagomir (ctrl antagomir) or antagomir specific for miR-92a (antagomir-92a), n refers to number of hearts. (f) Western blot analysis of endogenous Hand2 or GAPDH protein expression in hearts from mice treated with ctrl antagomir or antagomir-92a. (g) Quantification of GAPDH corrected Hand2 Western blot signals from (f), n refers to number of hearts. (h) Real-time PCR analysis of Hand2 transcript abundance in hearts from mice treated with ctrl antagomir or antagomir-92a, n refers to number of hearts. (i) Schematic representation of the precursor sequence of hsa-miR-25 and conservation level of the mature miR-25-3p strand. (j) Endogenous miR-25 expression in human fetal myocardium and human adult myocardium indicating a higher expression level of miR-25 in the adult human heart, n refers to number of hearts. (k) Northern blot analysis of miR-25 expression in multiple mouse organs. (l) Activity assay of luciferase reporter construct harboring an intact or mutated Hand2 3′UTR after transfection with premiR- 25, pre-miR-92a, pre-miR-92b or pre-miR-1 in neonatal rat cardiomyocytes. A scrambled precursor miR (scr.pre-miR) was used as a control, n refers to number of transfection experiments. (m) Confocal microscopy images of neonatal rat cardiomyocytes transfected with scrambled anti-miR (scr.anti-miR) or anti-miR for miR-25 (anti-miR-25) and treated with PE for 24 h. Quantification of cell surface areas from these conditions (lower panel), n refers to number of microscopic fields. *P<0.05 vs control group (error bars are s.e.m.). Source data are shown in Supplementary Table 9.
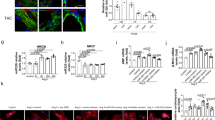
Supplementary Figure 5 Downregulation of the miR-106b∼25 cluster in the adult heart involves NFAT-dependent transcriptional repression.
(a) Confocal microscopy images of neonatal rat cardiomyocytes transfected with scrambled precursor (scr.pre-miR), synthetic precursor scrambled anti-miR (scr.anti-miR) or anti-miR for miR-92a (pre-miR-92b, anti-miR-92b), miR-92b (pre-miR-92b, anti-miR-92b) and treated with or without PE for 24 h. Scale bar, 50 μm. (b) Quantification of cell surface areas from conditions in (a), n refers to number of microscopic fields. (c) Confocal microscopy images of neonatal rat cardiomyocytes infected with AdLacz (control adenovirus), AdVIVIT (adenovirus overexpressing the VIVIT optimized NFAT inhibitory peptide), AdVIVIT + pre-miR-25 and AdHand2 + pre-miR-25. In all conditions cells were treated with PE or AdCnA for 24h. Scale bar, 50 μm. (d) Quantification of cell surface areas from conditions in (c), n refers to number of microscopic fields. (e) Pri-miR-25, pri-miR-106b and pri-miR-93 are down-regulated in two animal models of heart failure: TAC hearts and MHC-CnA Tg hearts, n refers to number of hearts. (f) Pre-miR-25, pre-miR-106b and pre-miR-93 are downregulated in two animal models of heart failure: TAC hearts and MHC-CnA Tg hearts, n refers to number of hearts. (g) Real-time PCR analysis of the MCM7-001 and MCM7-010 transcripts in wild-type (wt) versus MHC-CnA Tg hearts, n refers to number of hearts. (h) Real-time PCR analysis of miR-25 expression of neonatal rat cardiomyocytes infected with AdLacz, AdCnA, AdVIVIT, and AdCnA with AdVIVIT, n refers to number of dishes. (i) Real-time PCR analysis of MCM7-001 and MCM7-010 expression of neonatal rat cardiomyocytes infected with AdLacz, AdCnA, AdVIVIT, and AdCnA with AdVIVIT, n refers to number of dishes. *P<0.05 vs control group; #P<0.05 vs experimental group (error bars are s.e.m.). Source data are shown in Supplementary Table 9.
Supplementary Figure 6 Specific miR-25 silencing with antagomir-25.
(a) Real-time PCR analysis of miR-93 expression in hearts from mice receiving control (ctrl) antagomir or antagomir against miR-25 (antagomir-25) and subjected to sham or transverse aortic constriction (TAC) surgery, n refers to number of hearts. (b) Real-time PCR analysis of miR-92b expression in hearts from mice receiving control (ctrl) antagomir or antagomir against miR-25 and subjected to sham or transverse aortic constriction (TAC) surgery, n refers to number of hearts. (c) Real-time PCR analysis of miR-148a expression in hearts from mice receiving control (ctrl) antagomir or antagomir against miR-25 and subjected to sham or transverse aortic constriction (TAC) surgery, n refers to number of hearts. (d) Real-time PCR analysis of relative transcript abundance for the fetal marker genes Nppa, Nppb, Acta1 and Myh7 in hearts from mice treated with ctrl antagomir or antagomir-25 following sham or TAC surgery, n refers to number of hearts. *P<0.05 vs control group (error bars are s.e.m.). Source data are shown in Supplementary Table 9.
Supplementary information
Supplementary Information
Supplementary Information (PDF 1055 kb)
Supplementary Table 1
Supplementary Information (XLSX 43 kb)
Supplementary Table 2
Supplementary Information (XLSX 43 kb)
Supplementary Table 3
Supplementary Information (XLSX 41 kb)
Supplementary Table 4
Supplementary Information (XLSX 354 kb)
Supplementary Table 5
Supplementary Information (XLSX 141 kb)
Supplementary Table 6
Supplementary Information (XLSX 43 kb)
Supplementary Table 7
Supplementary Information (XLSX 45 kb)
Supplementary Table 8
Supplementary Information (XLSX 47 kb)
Supplementary Table 9
Supplementary Information (XLSX 99 kb)
Rights and permissions
About this article
Cite this article
Dirkx, E., Gladka, M., Philippen, L. et al. Nfat and miR-25 cooperate to reactivate the transcription factor Hand2 in heart failure. Nat Cell Biol 15, 1282–1293 (2013). https://doi.org/10.1038/ncb2866
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb2866
This article is cited by
-
Tuning the Stability of the Polyplex Nanovesicles of Oligonucleotides via a Zinc (II)-Coordinative Strategy
Chinese Journal of Polymer Science (2022)
-
Dickkopf 3: a Novel Target Gene of miR-25-3p in Promoting Fibrosis-Related Gene Expression in Myocardial Fibrosis
Journal of Cardiovascular Translational Research (2021)
-
A microRNA program regulates the balance between cardiomyocyte hyperplasia and hypertrophy and stimulates cardiac regeneration
Nature Communications (2021)
-
The role of dihydrosphingolipids in disease
Cellular and Molecular Life Sciences (2019)
-
LncRNA NRON alleviates atrial fibrosis via promoting NFATc3 phosphorylation
Molecular and Cellular Biochemistry (2019)