Abstract
TAp73 is a structural homologue of the pre-eminent tumour suppressor p53. However, unlike p53, TAp73 is rarely mutated, and instead is frequently overexpressed in human tumours. It remains unclear whether TAp73 affords an advantage to tumour cells and if so, what the underlying mechanism is. Here we show that TAp73 supports the proliferation of human and mouse tumour cells. TAp73 activates the expression of glucose-6-phosphate dehydrogenase (G6PD), the rate-limiting enzyme of the pentose phosphate pathway (PPP). By stimulating G6PD, TAp73 increases PPP flux and directs glucose to the production of NADPH and ribose, for the synthesis of macromolecules and detoxification of reactive oxygen species (ROS). The growth defect of TAp73-deficient cells can be rescued by either enforced G6PD expression or the presence of nucleosides plus an ROS scavenger. These findings establish a critical role for TAp73 in regulating metabolism, and connect TAp73 and the PPP to oncogenic cell growth.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Kaghad, M. et al. Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell 90, 809–819 (1997).
Vogelstein, B., Lane, D. & Levine, A. J. Surfing the p53 network. Nature 408, 307–310 (2000).
Vousden, K. H. & Prives, C. Blinded by the light: the growing complexity of p53. Cell 137, 413–431 (2009).
Melino, G., De Laurenzi, V. & Vousden, K. H. p73: friend or foe in tumorigenesis. Nat. Rev. Cancer 2, 605–615 (2002).
Yang, A., Kaghad, M., Caput, D. & McKeon, F. On the shoulders of giants: p63, p73 and the rise of p53. Trends Genet. 18, 90–95 (2002).
Moll, U. M. & Slade, N. p63 and p73: roles in development and tumour formation. Mol. Cancer Res. 2, 371–386 (2004).
Deyoung, M. P. & Ellisen, L. W. p63 and p73 in human cancer: defining the network. Oncogene 26, 5169–5183 (2007).
Donehower, L. A. et al. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature 356, 215–221 (1992).
Jacks, T. et al. Tumour spectrum analysis in p53-mutant mice. Curr. Biol. 4, 1–7 (1994).
Yang, A. et al. p73-deficient mice have neurological, pheromonal and inflammatory defects but lack spontaneous tumours. Nature 404, 99–103 (2000).
Flores, E. R. et al. Tumour predisposition in mice mutant for p63 and p73: evidence for broader tumour suppressor functions for the p53 family. Cancer Cell 7, 363–373 (2005).
Flores, E. R. et al. p63 and p73 are required for p53-dependent apoptosis in response to DNA damage. Nature 416, 560–564 (2002).
Senoo, M., Manis, J. P., Alt, F. W. & McKeon, F. p63 and p73 are not required for the development and p53-dependent apoptosis of T cells. Cancer Cell 6, 85–89 (2004).
Tomasini, R. et al. TAp73 knockout shows genomic instability with infertility and tumour suppressor functions. Genes Dev. 22, 2677–2691 (2008).
Talos, F., Nemajerova, A., Flores, E. R., Petrenko, O. & Moll, U. M. p73 suppresses polyploidy and aneuploidy in the absence of functional p53. Mol. Cell 27, 647–659 (2007).
Wilhelm, M. T. et al. Isoform-specific p73 knockout mice reveal a novel role for ΔNp73 in the DNA damage response pathway. Genes Dev. 24, 549–560 (2010).
Vander Heiden, M. G., Cantley, L. C. & Thompson, C. B. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029–1033 (2009).
Cairns, R. A., Harris, I. S. & Mak, T. W. Regulation of cancer cell metabolism. Nat. Rev. Cancer 11, 85–95 (2011).
Dang, C. V. Links between metabolism and cancer. Genes Dev. 26, 877–890 (2012).
Wellen, K. E. & Thompson, C. B. Cellular metabolic stress: considering how cells respond to nutrient excess. Mol. Cell 40, 323–332 (2010).
Levine, A. J. & Puzio-Kuter, A. M. The control of the metabolic switch in cancers by oncogenes and tumour suppressor genes. Science 330, 1340–1344 (2010).
Weinberg, F. et al. Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proc. Natl Acad. Sci. USA 107, 8788–8793 (2010).
Sena, L. A. & Chandel, N. S. Physiological roles of mitochondrial reactive oxygen species. Mol. Cell 48, 158–167 (2012).
Jiang, P. et al. p53 regulates biosynthesis through direct inactivation of glucose-6-phosphate dehydrogenase. Nat. Cell Biol. 13, 310–316 (2011).
Mancuso, A., Sharfstein, S. T., Tucker, S. N., Clark, D. S. & Blanch, H. W. Examination of primary metabolic pathways in a murine hybridoma with carbon-13 nuclear magnetic resonance spectroscopy. Biotechnol. Bioeng. 44, 563–585 (1994).
Berg, J. M., Tymoczko, J. L. & Stryer, L. Biochemistry 6th edn 577–589 (W. H. Freeman, 2006).
Gong, J. G. et al. The tyrosine kinase c-Abl regulates p73 in apoptotic response to cisplatin-induced DNA damage. Nature 399, 806–809 (1999).
Agami, R., Blandino, G., Oren, M. & Shaul, Y. Interaction of c-Abl and p73α and their collaboration to induce apoptosis. Nature 399, 809–813 (1999).
Yuan, Z. M. et al. p73 is regulated by tyrosine kinase c-Abl in the apoptotic response to DNA damage. Nature 399, 814–817 (1999).
Urist, M., Tanaka, T., Poyurovsky, M. V. & Prives, C. p73 induction after DNA damage is regulated by checkpoint kinases Chk1 and Chk2. Genes Dev. 18, 3041–3054 (2004).
Riley, T., Sontag, E., Chen, P. & Levine, A. Transcriptional control of human p53-regulated genes. Nat. Rev. Mol. Cell Biol. 9, 402–412 (2008).
Jost, C. A., Marin, M. C. & Kaelin, W. G. Jr p73 is a simian [correction of human] p53-related protein that can induce apoptosis. Nature 389, 191–194 (1997).
De Laurenzi, V. et al. Two new p73 splice variants, γ and δ, with different transcriptional activity. J. Exp. Med. 188, 1763–1768 (1998).
Di Como, C. J., Gaiddon, C. & Prives, C. p73 function is inhibited by tumour-derived p53 mutants in mammalian cells. Mol. Cell Biol. 19, 1438–1449 (1999).
Temple, M. D., Perrone, G. G. & Dawes, I. W. Complex cellular responses to reactive oxygen species. Trends Cell Biol. 15, 319–326 (2005).
Pandolfi, P. P. et al. Targeted disruption of the housekeeping gene encoding glucose 6-phosphate dehydrogenase (G6PD): G6PD is dispensable for pentose synthesis but essential for defense against oxidative stress. EMBO J. 14, 5209–5215 (1995).
Filosa, S. et al. Failure to increase glucose consumption through the pentose-phosphate pathway results in the death of glucose-6-phosphate dehydrogenase gene-deleted mouse embryonic stem cells subjected to oxidative stress. Biochem. J. 370, 935–943 (2003).
Rufini, A. et al. TAp73 depletion accelerates aging through metabolic dysregulation. Genes Dev. 26, 2009–2014 (2012).
Dworkin, C. R., Gorman, S. D., Pashko, L. L., Cristofalo, V. J. & Schwartz, A. G. Inhibition of growth of HeLa and WI-38 cells by dehydroepiandrosterone and its reversal by ribo- and deoxyribonucleosides. Life Sci. 38, 1451–1457 (1986).
Tian, W. N. et al. Importance of glucose-6-phosphate dehydrogenase activity for cell growth. J. Biol. Chem. 273, 10609–10617 (1998).
Son, J. et al. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature 496, 101–105 (2013).
Jiang, P., Du, W., Mancuso, A., Wellen, K. E. & Yang, X. Reciprocal regulation of p53 and malic enzymes modulates metabolism and senescence. Nature 493, 689–693 (2013).
Anastasiou, D. et al. Inhibition of pyruvate kinase M2 by reactive oxygenspecies contributes to cellular antioxidant responses. Science 334, 1278–1283 (2011).
Christofk, H. R., Vander Heiden, M. G., Wu, N., Asara, J. M. & Cantley, L. C. Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature 452, 181–186 (2008).
Li, Y., Zhou, Z. & Chen, C. WW domain-containing E3 ubiquitin protein ligase 1 targets p63 transcription factor for ubiquitin-mediated proteasomal degradation and regulates apoptosis. Cell Death Differ. 15, 1941–1951 (2008).
Brummelkamp, T. R., Bernards, R. & Agami, R. A system for stable expression of short interfering RNAs in mammalian cells. Science 296, 550–553 (2002).
Rocco, J. W., Leong, C. O., Kuperwasser, N., DeYoung, M. P. & Ellisen, L. W. p63 mediates survival in squamous cell carcinoma by suppression of p73-dependent apoptosis. Cancer Cell 9, 45–56 (2006).
Godar, S. et al. Growth-inhibitory and tumour- suppressive functions of p53 depend on its repression of CD44 expression. Cell 134, 62–73 (2008).
Leopold, J. A. et al. Aldosterone impairs vascular reactivity by decreasing glucose-6-phosphate dehydrogenase activity. Nat. Med. 13, 189–197 (2007).
Cossarizza, A. et al. Simultaneous analysis of reactive oxygen species and reduced glutathione content in living cells by polychromatic flow cytometry. Nat. Protoc. 4, 1790–1797 (2009).
Mancuso, A. et al. Real-time detection of 13C NMR labeling kinetics in perfused EMT6 mouse mammary tumour cells and betaHC9 mouse insulinomas. Biotechnol. Bioeng. 87, 835–848 (2004).
Bunz, F. et al. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science 282, 1497–1501 (1998).
Acknowledgements
We thank G. Melino (University Rome Tor Vergata, Rome, Italy), C. Chen (Albany Medical College, Albany, USA), A. Tullo (University of Rome La Sapienza, Rome, Italy), U. M. Moll (Stony Brook University, Stony Brook, New York, USA), M. C. Marín (Harvard Medical School, Boston, USA), B. Vogelstein (Johns Hopkins University, Baltimore, USA), W. El-Deiry (Penn State Hershey Cancer Institute, Pennsylvania, USA) for expression plasmids, antibodies and/or cell lines; W. Wang, Y. Mei, N. Li, X. Wang, W. Tan, E. Witze and K. E. Wellen for technical assistance; and C. O’Neill for help with manuscript preparation. Supported by China National Natural Science Foundation (31030046), Chinese Academy of Sciences (XDA01020104), Ministry of Science and Technology of China (2010CB912804 and 2011CB966302) and the Ministry of Education of China (20123402130006) to M.W., and US National Institutes of Health (CA088868 and GM060911) and the Department of Defense (W81XWH-10-1-0468) to X.Y.
Author information
Authors and Affiliations
Contributions
W.D., P.J., X.Y. and M.W. designed the experiments and interpreted results. W.D. and P.J. performed all the experiments except those mentioned below. A.M. and P.J. analysed the oxidative PPP flux. A.S. and M.D.B. helped with the FACS analysis and xenograft study, respectively. A.J.M. performed the breast cancer data analysis. T.W.M. supplied the MEF cells deficient for TAp73 and ΔNp73. W.D., P.J. and X.Y. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
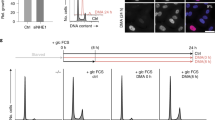
Supplementary Figure 1 Requirement of TAp73 for cell proliferation and tumor growth.
(a) Schematic representation of p53 and the TAp73 and ΔNp73 isoform classes. Each p73 isoform class comprises various splicing variants (α, β, γ etc.) that differ in their C-terminal regions. TA: transactivation domain. DBD: DNA-binding domain. OD: oligomerization domain. SAM: sterile alpha motif. (b) TAp73+/+ and TAp73−/− MEF cells cultured for 4 days were stained with crystal violet. (c) Representative images of animals two weeks after injected with TAp73+/+ and TAp73−/− MEF cells. (d) U2OS cells stably expressing control or TAp73 shRNA were stained with crystal violet at day 6. (e,f) Proliferation of two independent clones of ΔNp73−/− and ΔNp73+/+ MEFs in culture and images of crystal violet staining at day 6. Data are means ± SD (n = 3 independent experiments).
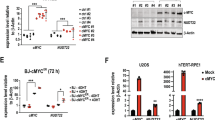
Supplementary Figure 2 p73 enhances G6PD expression.
(a) The pentose phosphate pathway and its link with glycolysis. FBP: fructose 1,6-biphosphate. PEP: phosphoenolpyruvate. TCA cycle: tricarboxylic acid cycle. (b) Expression of G6PD and TAp73 proteins in U2OS cells transfected with control siRNA or p73 siRNA, plus control vector or siRNA-resistant plasmid encoding Flag-TAp73. Western blots represent two independent experiments. (c) Semiquantitative RT-PCR analysis of ΔNp73 expression in HeLa, U2OS, H1299, and p53+/+ HCT116 cells. (d) Comparison of effect of p73 and ΔNp73 on G6PD expression in p73, ΔNp73 or control siRNA in HeLa cells by RT-PCR. (e) U2OS cells were infected with lentiviruses expressing a TAp73 shRNA or a control shRNA and cultured for the indicated time. The expression of G6PD, TAp73, and actin was analyzed by Western blot. The data represents three independent experiments. (f) mRNA expression in H1299 cells stably expressing control shRNA, p63 shRNA, or TAp73 shRNA was assayed by RT-PCR. (g) Quantitative RT-PCR analysis of G6PD expression in H1299 cells transfected with increasing amounts of control plasmid or plasmid expressing the indicated p53 family proteins. Data are means ± SD (n = 3 independent experiments). (h) A549 cells were treated with 1 μg/ml doxorubicin for 24 hour and protein expression was analyzed by Western blot. (i) Luciferase reporter assay of 293T cells transfected with p73 RE-luciferase construct or control construct, plus TAp73 or ΔNp73. Data are means ± SD (n = 3 independent experiments). (j) H1299 cells were transfected with increasing amounts of Flag-p53R175H, Flag-p53R273H, or Flag-TAp73α as indicated. Protein expression was examined by Western blot. The data represents two independent experiments.
Supplementary Figure 3 TAp73 regulates NADPH metabolism.
(a,b) NADPH levels (a) and relative NADP+/NADPH ratios (b) in ΔNp73+/+ and ΔNp73−/− MEF cells. Data are means ± SD (n = 3 independent experiments). (c) Relative NADPH levels in IMR90 cells transfected with control (−) or p73 siRNA. Data are means ± SD (n = 3 independent experiments). (d) NADP+/NADPH ratios of IMR90 cells treated with p73 or control siRNA. Data are means ± SD (n = 3 independent experiments). (e) Relative NADPH levels in p53+/+ and p53−/− HCT116 cells transfected with control (−) or p73 siRNA. Data are means ± SD (n = 3 independent experiments).
Supplementary Figure 4 A role of TAp73 in anti-oxidant defense.
(a) TAp73+/+ and TAp73−/− MEF cells treated with control or G6pd siRNA. ROS was measured by 2′, 7′-di-chlorofluorescein (DCF) staining and flow cytometry (left), and protein expression by Western blot (Right). Western blots represent three independent experiments. (b) ROS levels in TAp73+/+ and TAp73−/− MEF cells that were treated with or without DHEA. (c) IMR90 cells were transfected with p73, G6PD, and control siRNA as indicated. ROS was measured. (d) TAp73+/+ and TAp73−/− MEF cells were treated with or without 50 mM H2O2 for 30 min and then cultured for 24 h in the presence or absence of DHEA. Cell viability was analyzed. Data are means ± SD (n = 3 independent experiments). (e) U2OS cells were transfected with control siRNA (−), p73 siRNA, and G6PD siRNA as indicated. Cells were treated with or without 250 μM H2O2 for 24 h and cell viability was analyzed by trypan blue staining. Data are means ± SD (n = 3 independent experiments).
Supplementary Figure 5 p73 regulates DNA synthesis and cell senescence via G6PD.
(a,b) p53−/− and p21−/− HCT116 cells were transfected with control (−), p73, and G6PD siRNA as indicated. Cells were assayed for BrdU incorporation. Data are means ± SD (n = 3 independent experiments). (c,d) Cell-cycle profile of p53−/− (c) and p21−/− (d) HCT116 cells transfected with p73, G6PD, and control siRNA as indicated. Protein expression is shown below. Western blots represent three independent experiments. (e) Percentage of SA- β-gal positive cells in IMR90 culture that were transfected with control, p73, or G6PD siRNA, and cultured for 72 h. Protein expression is shown below. Data are means ± SD (n = 3 independent experiments). Western blots represent three independent experiments.
Supplementary Figure 6 p73 promotes cell proliferation through G6PD.
(a–c) Proliferation of p53+/+ (a), p53−/− (b), and p21−/− (c) HCT116 cells transfected with the control, G6PD, and p73 siRNAs as indicated. Data are means ± SD (n = 3 independent experiments). (d) Growth of U2OS cells transfected with control siRNA or p73 siRNA, plus control vector or Flag-G6PD. Data are means ± SD (n = 3 independent experiments). Protein expression is shown on the Right. Western blots represent three independent experiments. (e–h) Growth of TAp73+/+ and TAp73−/− MEF cells stably overexpressing G6PD or vector control. Cells were cultured in medium containing vehicle (e), NAC (f), four ribonucleosides and four deoxyribonucleosides (Nuc) (g), or both NAC and Nuc (h). Data are means ± SD (n = 3 independent experiments).
Supplementary Figure 7 G6PD levels are associated with breast cancer metastasis and a role for G6PD and TAp73 in tumor growth.
(a,b) G6PD levels are associated with breast cancer metastasis. (a) Cox multivariable regression for metastasis risk using the indicated variables was performed on a cohort of 871 breast cancer patients. Shown are the hazard ratio (HR) and p-values. (b) Kaplan-Meier survival curves for metastasis-free survival. The patients were stratified by mean G6PD levels (‘hi’ is greater than mean, ‘lo’ is less than or equal to mean). P-value was determined by the log-rank test. (c) Representative images of animals three weeks after injected with TAp73+/+ and TAp73−/− MEF cells stably overexpressing G6PD or vector control. (d) Mice were injected with TAp73+/+ and TAp73−/− MEF cells that were treated with control or G6pd siRNA, and were fed with water containing no NAC or 40 mM NAC. Representative images of animals at three weeks are shown.
Supplementary information
Supplementary Information
Supplementary Information (PDF 1888 kb)
Rights and permissions
About this article
Cite this article
Du, W., Jiang, P., Mancuso, A. et al. TAp73 enhances the pentose phosphate pathway and supports cell proliferation. Nat Cell Biol 15, 991–1000 (2013). https://doi.org/10.1038/ncb2789
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb2789
This article is cited by
-
Metabolic reprogramming in colorectal cancer: regulatory networks and therapy
Cell & Bioscience (2023)
-
A critical bioenergetic switch is regulated by IGF2 during murine cartilage development
Communications Biology (2022)
-
The p53 family member p73 in the regulation of cell stress response
Biology Direct (2021)
-
NADPH levels affect cellular epigenetic state by inhibiting HDAC3–Ncor complex
Nature Metabolism (2021)
-
NF-κB and pSTAT3 synergistically drive G6PD overexpression and facilitate sensitivity to G6PD inhibition in ccRCC
Cancer Cell International (2020)